Dihydrolipoamide acetyltransferase
From Proteopedia
(Difference between revisions)
| (14 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
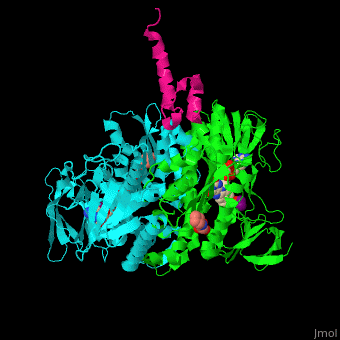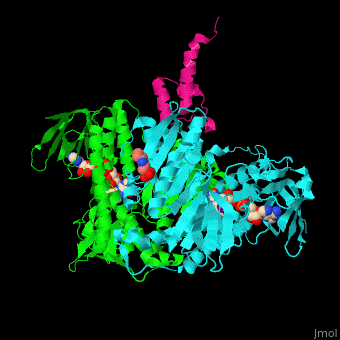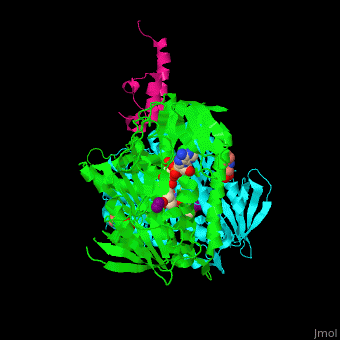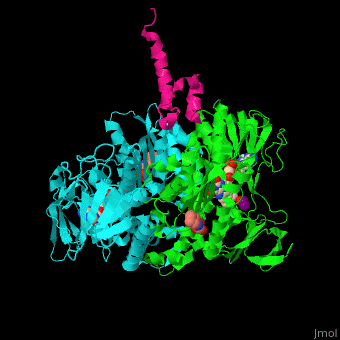
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='Human dihydrolipoamide acetyltransferase binding domain (cyan, green) complex with E3 binding domain (magenta), FAD, CHES and mercaptodthanol (PDB entry [[3rnm]])' scene='48/485632/Cv/3'> |
| - | '''Dihydrolipoamide acetyltransferase''' (DLAT) or E2 is part of the pyruvate dehydrogenase complex together with pyruvate dehydrogenase (E1) and dihydrolipoyl dehydrogenase (E3). The complex decarboxylates pyruvate thus linking the glycolysis to the citric acid cycle. DLAT catalyzes the transfer of acetyl group to CoA.<ref>PMID:14736882</ref> | + | '''Dihydrolipoamide acetyltransferase''' (DLAT) or E2 is part of the pyruvate dehydrogenase complex together with pyruvate dehydrogenase (E1) and dihydrolipoyl dehydrogenase (E3). The complex decarboxylates pyruvate thus linking the glycolysis to the citric acid cycle. DLAT catalyzes the transfer of acetyl group to CoA.<ref>PMID:14736882</ref> |
| - | = | + | <scene name='48/485632/Cv/9'>Human dihydrolipoamide acetyltransferase binding domain with E3 binding domain, FAD, CHES and mercaptodthanol</scene>. |
| - | <scene name='48/485632/Cv/ | + | <scene name='48/485632/Cv/10'>FAD binding site</scene>. |
| + | |||
| + | <scene name='48/485632/Cv/11'>Mercaptodthanol binding site</scene>. | ||
| + | |||
| + | <scene name='48/485632/Cv/12'>CHES binding site</scene> (PDB entry [[3rnm]]), water molecules shown as red spheres. | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structures of dihydrolipoamide acetyltransferase== | ==3D structures of dihydrolipoamide acetyltransferase== | ||
| + | [[Dihydrolipoamide acetyltransferase 3D structures]] | ||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *DLAT lipoyl domain | ||
| - | |||
| - | **[[1lab]], [[1lac]] – GsDLAT lipoyl domain – ''Geobacillus stearothermophilus'' – NMR<BR /> | ||
| - | **[[1iyu]], [[1iyv]] - DLAT lipoyl domain – ''Azotobacter vinelandii'' – NMR<BR /> | ||
| - | **[[1fyc]] - DLAT lipoyl domain – human – NMR<BR /> | ||
| - | **[[1qjo]] - EcDLAT lipoyl domain – ''Escherichia coli'' – NMR<BR /> | ||
| - | |||
| - | *DLAT binding domain | ||
| - | |||
| - | **[[2pdd]], [[2pde]] – GsDLAT binding domain – NMR<BR /> | ||
| - | **[[1w4e]], [[1w4f]], [[1w4g]] - GsDLAT binding domain (mutant) – NMR<BR /> | ||
| - | **[[1w4h]] - EcDLAT binding domain – NMR<BR /> | ||
| - | **[[1w4i]] - PaDLAT binding domain – ''Pyrobaculum aerophilum'' – NMR<BR /> | ||
| - | **[[1w4j]], [[1w4k]] - PaDLAT binding domain (mutant) – NMR<BR /> | ||
| - | **[[1ebd]], [[2eq8]], [[2eq9]] - GsDLAT binding domain + E3<BR /> | ||
| - | **[[3rnm]] - hDLAT binding domain (mutant) + E3<BR /> | ||
| - | **[[1w85]], [[1w88]] - GsDLAT binding domain + E1<br /> | ||
| - | **[[1y8n]], [[1y8o]], [[1y8p]] - hDLAT binding domain + pyruvate dehydrogenase kinase | ||
| - | |||
| - | *DLAT catalytic domain | ||
| - | |||
| - | **[[1b5s]] - GsDLAT catalytic domain | ||
| - | |||
| - | **[[2k7v]] – GsDLAT residues 206-293 – NMR<BR /> | ||
| - | |||
| - | *Full DLAT | ||
| - | |||
| - | **[[3duf]], [[3dv0]], [[3dva]] - GsDLAT + E1 | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of dihydrolipoamide acetyltransferase
Dihydrolipoamide acetyltransferase 3D structures
References
- ↑ Lai WL, Chou LY, Ting CY, Kirby R, Tsai YC, Wang AH, Liaw SH. The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation. J Biol Chem. 2004 Apr 2;279(14):13962-7. Epub 2004 Jan 21. PMID:14736882 doi:http://dx.doi.org/10.1074/jbc.M308849200