DNA adenine methylase
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
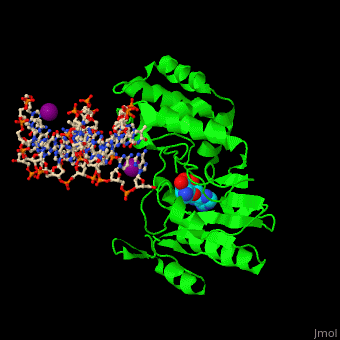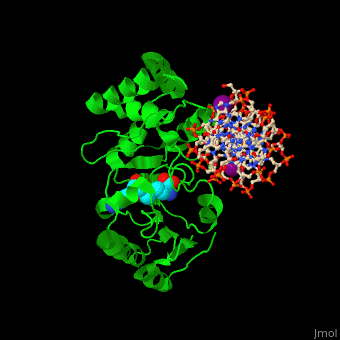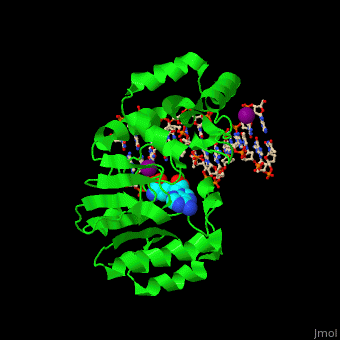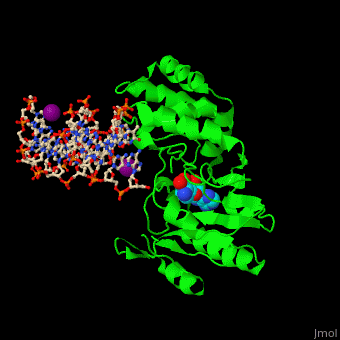
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='Phage T4 DNA adenine metylase complex with SAH, DNA and I- ion (purple) (PDB code [[1q0t]])' scene='51/517445/Cv/1'> |
| - | + | ||
| - | + | ||
== Function == | == Function == | ||
| - | + | '''DNA adenine methylase''' (DAM) adds a methyl group to adenines in the DNA sequence GATC. The daughter strand remains unmethylated for a short while during which mismatch repair of DNA can take place. DAM has a role in regulation of replication by delaying the availability of the origin of replication to activation during the cell cycle. The origin of replication in ''E. coli'' contains 11 GATC sequences. In ''E. coli'', downstream GATC sequences are methylated to promote RNA transcription. <ref>PMID:15802246</ref> | |
| - | + | ||
| - | + | ||
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | DAM uses S-adenosyl-L-homocysteine (AdoHcy, SAH) as methyl donor. | ||
| + | |||
| + | <scene name='51/517445/Cv/10'>Phage T4 DNA adenine metylase with SAH, DNA and I- ions</scene> | ||
| + | |||
| + | <scene name='51/517445/Cv/11'>SAH binding site</scene> | ||
| + | |||
| + | <scene name='51/517445/Cv/12'>DNA binding site</scene> | ||
| + | |||
| + | <scene name='51/517445/Cv/13'>Iod coordination site</scene> (PDB code [[1q0t]]) <ref>PMID:12937411</ref> | ||
</StructureSection> | </StructureSection> | ||
| Line 19: | Line 25: | ||
[[1q0t]], [[1yf3]], [[1yfj]] – T4DAM + SAH + DNA <BR /> | [[1q0t]], [[1yf3]], [[1yfj]] – T4DAM + SAH + DNA <BR /> | ||
[[1yfl]] – T4DAM + sinefungin + DNA <BR /> | [[1yfl]] – T4DAM + sinefungin + DNA <BR /> | ||
| - | [[2g1p]], [[4rtk]], [[4rtn]], [[4rtq]] – EcDAM + SAH + DNA – Escherichia coli<BR /> | + | [[2g1p]], [[4rtk]], [[4rtn]], [[4rtq]] – EcDAM + SAH + DNA – ''Escherichia coli''<BR /> |
[[2ore]], [[4gbe]] – EcDAM + SAH <BR /> | [[2ore]], [[4gbe]] – EcDAM + SAH <BR /> | ||
[[4gol]], [[4gom]], [[4gon]], [[4goo]] – EcDAM + Aza-SAM derivative<BR /> | [[4gol]], [[4gom]], [[4gon]], [[4goo]] – EcDAM + Aza-SAM derivative<BR /> | ||
| Line 27: | Line 33: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D Structures of DNA adenine methylase
Updated on 25-February-2019
1q0s – T4DAM + SAH – Enterobacter phage T4
1q0t, 1yf3, 1yfj – T4DAM + SAH + DNA
1yfl – T4DAM + sinefungin + DNA
2g1p, 4rtk, 4rtn, 4rtq – EcDAM + SAH + DNA – Escherichia coli
2ore, 4gbe – EcDAM + SAH
4gol, 4gom, 4gon, 4goo – EcDAM + Aza-SAM derivative
4rtj, 4rtl, 4rto – EcDAM + sinefungin + DNA
4rtm, 4rtp, 4rtr, 4rts – EcDAM + AdoMet + DNA
References
- ↑ Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005 Apr;8(2):154-60. PMID:15802246 doi:http://dx.doi.org/10.1016/j.mib.2005.02.009
- ↑ Yang Z, Horton JR, Zhou L, Zhang XJ, Dong A, Zhang X, Schlagman SL, Kossykh V, Hattman S, Cheng X. Structure of the bacteriophage T4 DNA adenine methyltransferase. Nat Struct Biol. 2003 Oct;10(10):849-55. Epub 2003 Aug 24. PMID:12937411 doi:http://dx.doi.org/10.1038/nsb973