Sandbox Reserved 1160
From Proteopedia
(Difference between revisions)
(New page: {{Sandbox_Reserved_CH462_Central_Metabolism}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' s...) |
|||
| (119 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | == Human metabotropic glutamate receptor 5 transmembrane domain == | |
| - | == | + | <StructureSection load='4oo9' size='300' frame='true' side='right' caption='Human metabotropic glutamate receptor 5 transmembrane domain bound to mavoglurant (PDB code of [http://www.rcsb.org/pdb/explore/explore.do?structureId=4oo9 4oo9]). The 7 helices comprise the bulk of the protein structure. mGlu5 receptor is an important part of the glutamate signaling pathway ' scene='72/721531/Protien_lys/3'> |
| - | <StructureSection load=' | + | Receiving and responding to extracellular messages is critical to the proper function of the nervous system. Glutamate is the primary excitory neurotransmitter of the Central Nervous System (CNS), and metabotropic glutamate receptor 5 is a key member of the glutamate signaling pathway<ref name="Dore" />. Metabotropic glutamate receptor 5 is a homodimeric [[GPCR]] that resides in the cellular membrane <ref name="Dore" />. mGlu5 is a member of the Class C GPCR family and can further be categorized into the Group I subgroup<ref name="Wu" />. |
| - | + | The functionality of the mGlu5 receptor is determined by conformational changes throughout multiple domains. mGlu5 will bind glutamate through its extracellular Venus flytrap domain and the signal will be transduced across the membrane to a heterotrimeric G protein, which will ultimately lead to calcium release and the activation of [[PKC]]<ref name="Wu" />. The signal is relayed through a Gq/11 pathway<ref name="Dore" />. Activated PKC will elicit a excitory post-synaptic response and modulate long term potentiation in the CNS<ref name="Wu" />. | |
| - | + | ||
| - | == | + | Human mGlu5 is found throughout the central nervous system. Areas containing high concentrations of mGlu5 are often involved in emotional processing and higher cognition<ref name="Niswender" />. The localization of mGlu5 in the CNS and the presence of multiple domains makes mGlu5 an excellent target for treating neurological conditions including: schizophrenia, [http://www.fragilex.org/fragile-x/fragile-x-syndrome/ Fragile X], [http://www.en.wikipedia.org/wiki/Depression_(mood)depression], [http://www.en.wikipedia.org/wiki/Anxiety_disorder anxiety], and [http://www.en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease]<ref name="Wu" />. |
| - | == | + | == Discovery == |
| + | The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | ||
| - | == | + | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The structure of the <font color='darkgreen'>'''alpha helices'''</font> are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator <span style="color:yellow;background-color:black;font-weight:bold;">mavoglurant</span> shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 -<scene name='72/721531/Protien_lys/3'>Lysozyme</scene> was inserted into intracellular loop 2 to add stability<ref name="Dore" />. |
| + | == Structure== | ||
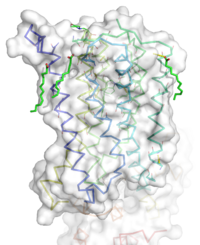
| + | [[Image:STR.png|200 px|left|thumb|'''Figure 1''': Overall Structure of the mGlu5 TMD. The polar heads on the Oleic acids orient the image with the top oriented extracellularly, the middle portion inserted into the membrane, and the lower portion oriented intracellularly. The white exterior represents the surface of the protien, and the multicolored lines interior to the surface represent the backbones 7 transmembrane alpha helices. ]] | ||
| + | === Overview === | ||
| + | The mGlu5 TMD contains 7 <scene name='72/721531/Protien_7_helices/4'> alpha helices</scene> that spans the membrane. The protein was crystallized with <scene name='72/721531/Protien_clean_sce/6'>Oleic acid</scene> shown in <font color='red'>'''red'''</font>. On the top portion of the protein several critical extracellular loops connect the TMD to the CRD. The binding pocket can be found deep in the interior of the protein, and is mainly comprised of hydrophobic amino acids with more polar amino acids found in the upper and lower portions of the binding site<ref name="Dore" />. Inserted into the biding pocket is the negative allosteric modulator [http://www.en.wikipedia.org/wiki/Mavoglurant mavoglurant]. The TMD is in an inactive conformation, since mavoglurant is bound<ref name="Dore" />. Also, the deletion of the flexible domains leaves the mGlu5 receptor unable to bind to its [[GPCR]]<ref name="Dore" />. The inactive state is maintained by multiple ionic locks whose positions determine the active versus inactive conformation. | ||
| - | == | + | When mGlu5 is in the active conformation, signaling begins with glutamate binding to the Venus flytrap domain. The signal is transduced across the cysteine-rich domain to the TMD<ref name="Niswender" />. Next, the dimerization of the TMD occurs. This activates the Gq/11 pathway, which activates phospholipase Cβ<ref name="Niswender" />. The active [http://www.proteopedia.org/wiki/index.php/2zkm phospholipase Cβ] hydrolyzes phosphotinositides and generates [https://pubchem.ncbi.nlm.nih.gov/compound/439456#section=Top inositol 1,4,5-trisphosphate] and [http://www.sivabio.50webs.com/ip3.htm diacyl-glycerol]<ref name="Woodcock" />. This results in calcium mobilization and activation of protein kinase C ([[PKC]])<ref name="Niswender" />. Calcium is a neurotransmitter, and relatively low concentrations of calcium can cause a large response across the neuronal synapse<ref name="Niswender" />. In addition to calcium stimulating an excitory response in nerve cells, PKC can be activated for regulatory purposes by the influx of calcium. A serine on PKC can become phosphorylated, which leaves PKC unable to bind to G beta-gamma (Gßγ) protein complex<ref name="Niswender" />. Unbound Gßγ protein can then inhibit voltage-sensitive calcium channels to reduce calcium influx and provide feedback inhibition to the glutamate signaling pathway. <ref name="Niswender" />. |
| + | === Extracellular Domain === | ||
| + | The extracellular domain of mGlu5 contains several key extracellular loops that will help modulate ligand binding. <scene name='72/721532/Ecl_trail_7/2'>Extracellular Loops</scene> are shown here with extracellular loops (ECLs) <font color='purple'><b>1</b></font>, <font color='purple'><b>2</b></font>, and <font color='purple'><b>3</b></font> highlighted in <font color='purple'><b>purple</b></font>. Additionally in the ECL domain, a <scene name='72/721531/Ecl_trail_1/5'>disulfide bond</scene> is attached to both Helix 3 and the amino acid chain between Helix 5 and the N-terminus. <font color='teal'><b>Helix 3</b></font> and <font color='red'><b>Helix 5</b></font> are colored in <font color='teal'><b>teal</b></font> and <font color='red'><b>red</b></font> respectively.The <font color='blue'><b>N-terminus</b></font> is represented in <font color='blue'><b>blue</b></font>. The <span style="color:yellow;background-color:black;font-weight:bold;">disulfide bond</span> is highlighted in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>, and it is conserved in all classes of mGlu5 TMD<ref name="Wu" />. The disulfide bond is critical in maintaining the position of ECL 2 <ref name="Dore" />. ECLs and the helices are also factors that dictate how mavoglurant fits in the binding pocket <ref name="Dore" />. The position of these ECLs can change the effective size of the binding pocket through loop positioning<ref name="Dore" />. | ||
| + | === Binding Pocket === | ||
| + | [[Image: Organic with clipped surface.png|200 px|left|thumb|'''Figure 2.''' Mavoglurant in its binding pocket of the 7TM region of mGLu5 Class C receptor. The binding pocket's surface is clipped in black with the substrate, mavoglurant, in red. The rest of the protein is colored in green. The binding pocket is present in the 7 Helix Transmembrane Domain that would be present in the phospholipid bilayer as an integral protein. The presence of mavoglurant inhibits the function of the metabotropic glutamate receptor.]] | ||
| + | The binding pocket, not to be confused with the glutamate binding site, represents a useful source of regulatory control. The binding pocket is only accessible by a relatively narrow (7 Å) <scene name='72/721531/Bindingsite/2'>entrance</scene><ref name="Dore" />. This small entrance severely restricts the access of both positive and negative allosteric regulators. One such regulator, <scene name='72/721531/Mavo/2'>mavoglurant</scene>, is a relatively small molecule that can travel through the entrance and fit between the helices of the mGlu5 receptor<ref name="Dore" />. Mavoglurant acts as a negative allosteric modulator of the mGlu5 receptor<ref name="Dore" />. Mavoglurant is a relatively hydrophobic molecule, which compliments the largely hydrophobic nature of the interior of mGlu5<ref name="Dore" />. The carbamate tail and hydroxyl group of mavoglurant will also interact with several key amino acids in the binding site of mGlu5<ref name="Dore" />. | ||
| + | |||
| + | Important Amino Acids<ref name="Dore" />: | ||
| + | *<scene name='72/721531/Protien_bindtop/8'>Asparagine</scene> 747 forms a hydrogen bond network with the main chain carbonyl of Glycine 652 and the carbamate portion of mavoglurant. | ||
| + | *Bicyclic ring of mavoglurant surrounded by <scene name='72/721531/Protien_hydrophobic/1'>hydrophobic binding pocket</scene>. <font color='purple'>'''Hydrophobic regions'''</font> of the mGlu5 receptor are shown in <font color='purple'>'''purple'''</font> and the <font color='grey'>'''rest of the helices'''</font> are shown in <font color='grey'>'''grey'''</font>. Mavoglurant is represented by spheres. | ||
| + | *2 Catalytic <scene name='72/721531/Protien_bindmiddle/4'>serine</scene> residues H-bond to the hydroxyl oxygen of our ligand. | ||
| + | *A <scene name='72/721531/Protien_bindbottom/2'>water molecule</scene> inside of the binding pocket helps stabilize the inactive state. | ||
| + | |||
| + | Upon mavoglurant’s diffusion into the mGlu5 receptor binding pocket, the ordered water cage found in the center of mGlu5 is displaced<ref name="Dore" />. Favorable interactions between the hydrophobic regions of the binding pocket and the bicyclic ring and the 3-methylphenyl ring of mavoglurant help increase the strength and energetic favorability of mavoglurant binding via the hydrophobic effect. In the binding pocket mavoglurant can hydrogen bond with Asp 747 and that hydrogen bond will be further stabilized by an extended hydrogen bond network with Gly 652<ref name="Dore" />. The hydroxyl group of mavoglurant will also form another hydrogen bond with Ser 805 and Ser 809<ref name="Dore" />. Multiple hydrogen bonds make the binding of mavoglurant to the mGlu5 receptor favorable and specific<ref name="Wu" />. | ||
| + | |||
| + | Once bound to mavoglurant, transmembrane helix 7 undergoes a conformational change<ref name="Dore" />. The shifting of TM7 will lead to a more global conformational change, which inactivates the receptor by moving intracellular loops inward <ref name="Dore" />. This will leave the G-protien unable to bind with mGlu5<ref name="Wu" />. Variation can be seen in positioning of alpha helices 5 and 7 across receptor class. Class C receptors have less space for mavoglurant to enter as compared to Class A and F receptors<ref name="Wu" />. Increased specificity and stronger binding affinity could be a result of the more narrow structure of mGlu5. | ||
| + | |||
| + | === Ionic Locks === | ||
| + | Another important structural feature is the series of <scene name='72/721531/Ionic_lock/5'>2 ionic locks</scene> on the intracellular side of the protein. Interactions between these five amino acids will form a salt bridge, which will stabilize the inactive conformation<ref name="Dore" />. The primary ionic lock forms between Glu770, Lys665, and Ser613<ref name="Dore" />. A secondary ionic lock occurs between Ser614 and Arg668<ref name="Dore" />. The purpose of these ionic locks is analogous to the ionic interactions that stabilize the T state in [[Hemoglobin]]. In the case of the TMD of mGlu5, the ionic lock is formed when the NAM mavoglurant is bound. These <scene name='72/721531/Ionic_lock/5'>ionic locks</scene> stabilize the inactive state, where the intracellular loops are stabilized facing inwards<ref name="Wu" />. This conformational change will effectively block the crevice that is involved in binding the G-protein<ref name="Wu" />. Models have suggested that, even in a glutamate bound state, the mavoglurant bound receptor would be dimerized but incapable of signaling<ref name="Wu" />. This signaling incapable mGlu5 dimer will help maintain the readiness of the pathway, while still decreasing signal response. | ||
| + | |||
| + | == Disease == | ||
| + | === Fragile X === | ||
| + | Fragile X syndrome is the most common genetic cause of mental disability, and is a member of the [http://www.nimh.nih.gov/health/topics/autism-spectrum-disorders-asd/index.shtml Autism spectrum disorder] family<ref name="Bailey" />. The severity of intellectual disability can vary from patient to patient, but symptoms commonly stem from a misregulation of the mGlu1 and MGlu5 pathways<ref name="Bailey" />. Misregulation of these pathways leads to over potentiation in neural cells. <scene name='72/721531/Mavo/2'>Mavoglurant</scene> and other allosteric regulators like [http://www.en.wikipedia.org/wiki/Fenobam fenobam] have shown promise in treating Fragile X. One positive characteristic of ligands that target the TMD of mGlu5 is they tend to be more specific, thus interacting less with nonspecific brain proteins<ref name="Feng" />. Mavoglurant down regulates glutamate signaling in an attempt to decrease potentiation. Unfortunately, recent Phase 2 clinical trials have proven mavoglurant ineffective <ref name="Bailey" />. However, modulators of mGlu5 TMD are still being investigated as treatment for Parkinson's, Alzheimer's disease, and various addictions<ref name="Niswender" />. | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | </StructureSection> | ||
== References == | == References == | ||
| + | <ref name="Dore">PMID: 25042998</ref> | ||
| + | <ref name="Wu">PMID: 24603153</ref> | ||
| + | <ref name="Niswender">PMID: 20055706</ref> | ||
| + | <ref name="Feng">PMID: 25762450</ref> | ||
| + | <ref name="Bailey">PMID: 26855682</ref> | ||
| + | <ref name="Woodcock">PMID: 18940816</ref> | ||
<references/> | <references/> | ||
| + | == External Resources == | ||
| + | [http://www.fraxa.org/novartis-discontinues-development-mavoglurant-afq056-fragile-x-syndrome/ Novartis Fragile X trials] | ||
| + | |||
| + | [http://www.researchgate.net/publication/263354624_Mavoglurant_as_a_treatment_for_Parkinson's_disease Mavoglurant as a treatment for Parkinson's disease] | ||
| + | |||
| + | [http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=293 Pharmacology of mGlu5 receptors ] | ||
| + | |||
| + | [http://en.wikipedia.org/wiki/Glutamate_receptor Glutamate signaling pathway and receptors general overview] | ||
| + | |||
| + | [http://www.en.wikipedia.org/wiki/Gq_alpha_subunit Gq Alpha subunit] | ||
Current revision
Human metabotropic glutamate receptor 5 transmembrane domain
| |||||||||||