Sandbox Reserved 1181
From Proteopedia
(Difference between revisions)
(New page: {{Sandbox_Reserved_CH462_Central_Metabolism}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' s...) |
|||
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <Structure load='4L6R' size='350' frame='true' align='right' caption='7TM structure of human class B GPCR 4L6R' scene='72/727091/Full_structure_with_labels/1'/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | <scene name='72/727091/Full_Structure_with_Labels/1'>Labels</scene> |
| - | = | + | <scene name='72/721552/Ligand_binding_interactions/1'>Ligand Binding Interactions and Crucial Disulfide Bond</scene> |
| - | = | + | <scene name='72/721552/Glucagon_binding/3'>Glucagon Binding Full Rendering</scene> |
| - | = | + | <scene name='72/721552/Glucagon_binding_zoomed_in/1'>Glucagon Binding Zoomed in</scene> |
| - | + | Old Stuff: | |
| - | </ | + | [[Image:ALA135PRO stalk unwinding.png|150 px|left|thumb|Fig. 1: A135P Mutation and effect on stalk stability <ref name= "Siu 2013"/>. ]] [[Image:Gln142 Tyr138 Glucagon interaction.png|150 px|right|thumb|Fig. 2: Stalk stabilized by salt bridge between Glu133-Lys136. Residues in yellow are demonstrated to have an effect on ligand binding affinity.<ref name= "Siu 2013"/>]] |
| - | == | + | |
| - | < | + | [[Image:Asn_298__Trp_295.png|150 px|left|thumb|Fig. 3: Active sites linked to glucagon binding affinity located on ECL1 are labeled<ref name= "Siu 2013"/>.]] |
| + | |||
| + | [[Image:Corticotropin class B and Glucagon class B receptors aligned.png |150 px|left|thumb|Fig. 4: Corticotropin-releasing factor 1 and glucagon receptors; Class B GPCRs with notable central splay]] [[Image:Beta2 class A and Glucagon class B receptors aligned.png |150 px|right|thumb|Fig. 5: Beta 2-adrenergic (class A) and glucagon receptors; showing an absence of central splay in Class A GPCRs.]] | ||
| + | |||
| + | [[Image:Movie_Frame_2.png|100 px|left|thumb|Fig. 7: Active site buried deep in 7TMD of glucagon receptor.]] | ||
| + | |||
| + | [[Image:Movie_Frame_7.png|175 px|left|thumb|Fig. 14: Distance measurement of GCGR 7TMD Y138-D362 of 19-20 angstroms and labeled with complimentary glucagon interaction residues.]] | ||
| + | [[Image:H1___Y10_with_measurement.png|175 px|right|thumb|Fig. 15: Distance measurement of H1-Y10 of 22-24 angstroms and labeled with complimentary GCGR 7TMD residue interactions.]] | ||
| + | |||
| + | ===Future research direction=== | ||
| + | Research for Class A GPCRs is much more extensive than for its secretin, class B counterparts, although class B is proving to be a worthwhile to invest researching. The challenge of class B stabilization, expression, and molecular size , has made class B GPCRs particularly hard to assay. Biochemical research has increased in the class B specifications, because it has been realized that receptors can be modulated by more than the agonist and antagonists present in vivo. Leading research consists of a complex interwoven scheme of equilibria manipulation in multi-receptor conformations. <ref name="Salon 2011"/> | ||
Current revision
|
Old Stuff:
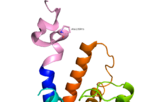
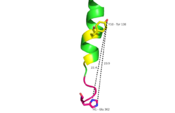
Fig. 1: A135P Mutation and effect on stalk stability [1].
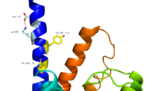
Fig. 2: Stalk stabilized by salt bridge between Glu133-Lys136. Residues in yellow are demonstrated to have an effect on ligand binding affinity.[1]
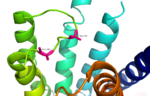
Fig. 3: Active sites linked to glucagon binding affinity located on ECL1 are labeled[1].
Future research direction
Research for Class A GPCRs is much more extensive than for its secretin, class B counterparts, although class B is proving to be a worthwhile to invest researching. The challenge of class B stabilization, expression, and molecular size , has made class B GPCRs particularly hard to assay. Biochemical research has increased in the class B specifications, because it has been realized that receptors can be modulated by more than the agonist and antagonists present in vivo. Leading research consists of a complex interwoven scheme of equilibria manipulation in multi-receptor conformations. [2]