We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Anum-II
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
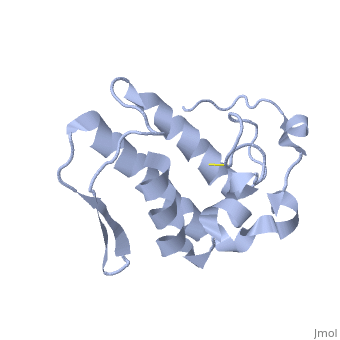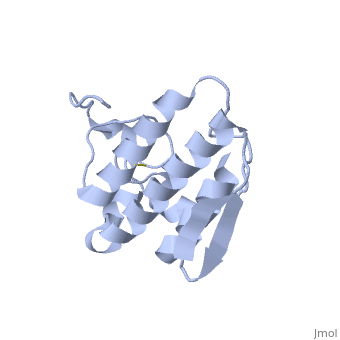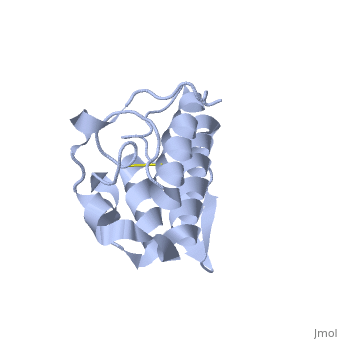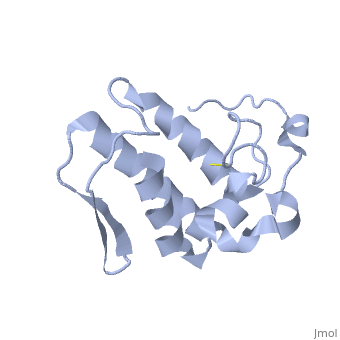
| - | Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel <scene name='Sandbox136/Feuillets/1'>sheet</scene> linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to <scene name='Sandbox136/Disulfide/1'>disulfide bonds </scene> ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a <scene name='Sandbox136/Stabilisation/1'>rigid platform</scene>. The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the <scene name='Sandbox136/Site_catalytique/2'>catalityc apparatus</scene> are conserved (His48,Tyr52, Tyr73 and Asp99) except for Asp49 which is replaced by Lys 49. | + | Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel <scene name='Sandbox136/Feuillets/1'>sheet</scene> linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to <scene name='Sandbox136/Disulfide/1'>disulfide bonds </scene> ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a <scene name='Sandbox136/Stabilisation/1'>rigid platform</scene>. The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the <scene name='Sandbox136/Site_catalytique/2'>catalityc apparatus</scene> are conserved (<scene name='49/497100/Active_site_residues_with_fas/2'>His48,Tyr52, Tyr73 and Asp99</scene>) except for Asp49 which is replaced by Lys 49. |
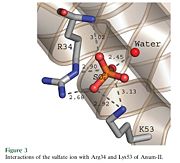
The structure of the protein has revealed the presence of an anion-binding site (Murakami ''et al.'', 2006) between <scene name='Sandbox136/Anion/2'>R34</scene>, <scene name='Sandbox136/Anion/2'>K53</scene> and a water molecule (see Figure 3). Sulfate ion is anchored thanks to hydrogen bonds between: | The structure of the protein has revealed the presence of an anion-binding site (Murakami ''et al.'', 2006) between <scene name='Sandbox136/Anion/2'>R34</scene>, <scene name='Sandbox136/Anion/2'>K53</scene> and a water molecule (see Figure 3). Sulfate ion is anchored thanks to hydrogen bonds between: | ||
Current revision
| |||||||||||
Bibliography
Holland, D.R., Clancy, L.L., Muchmore, S.W., Ryde, T.J., Einspahr, H.M., Finzel, B.C., Heinrikson, R.L., Watenpaugh, K.D. (1990).Biochem-J .265,17649-56
Chioato, L., De Oliveira, A. H., Ruller, R., Sa., J. M., Ward, R. J. (2002).Biochem-J .366,971-6
Murakami, M. T. , Melo,C. C., Angulo,Y. , Lomonte,B. Arni, R. K. (2006).Acta Cryst.F62, 423-426
Yamazaki, Y., Matsunaga, Y., Nakano, Y. & Morita, T. (2005). J. Biol. Chem. 280, 29989–29992.
Created with the participation of Elie Peillon