Phosphoglycerate Mutase
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
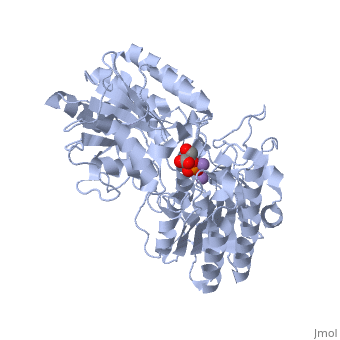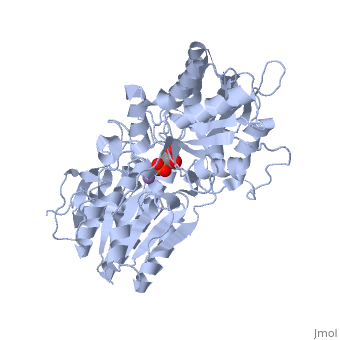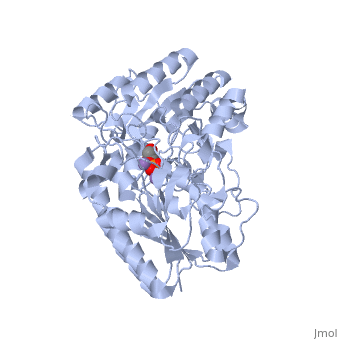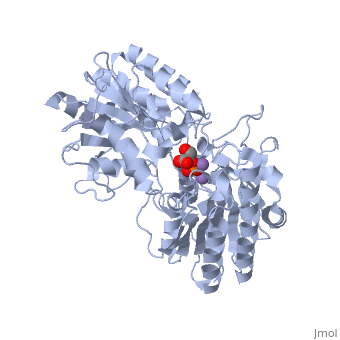
| - | + | <StructureSection load='1eqj' size='350' side='right' caption='Phosphoglycerate mutase complex with phosphoglyceric acid and Mn+2 ion (purple) [[1eqj]]' scene='' pspeed='8'> | |
== Background == | == Background == | ||
| Line 19: | Line 19: | ||
It is important to note that the phosphate group that is placed on C2 is not the same phosphate group that was initially on C3. | It is important to note that the phosphate group that is placed on C2 is not the same phosphate group that was initially on C3. | ||
| - | In order to understand how PGM catalyzes this reaction, an explanation of its active site is imperative. The most important residues in this enzyme include <scene name=' | + | In order to understand how PGM catalyzes this reaction, an explanation of its active site is imperative. The most important residues in this enzyme include <scene name='40/401494/Cv/1'>His 8 and 181</scene> with imidazole groups which are in close proximity to carbons 2 and 3 in the substrate. His-8 is phosphorylated during during catalysis, and it is likely that His-179 acts as the proton donor/acceptor <ref>Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253</ref>. Based on crystallography experiments, the active site where these histidine residues reside lies at the bottom of a deep groove in each subunit. <ref name="winn" /> The sites in each subunit, whether the enzyme is a homodimer or homotetramer, are well separated. The active enzyme contains a phosphoryl group attached to His 8. This phosphoryl group is what is transferred to C2 of the substrate, resulting in an intermediate 2,3-bisphosphoglycerate-enzyme complex. Thus there is a <scene name='40/401494/Cv/3'>covalently attached phosphate</scene> in the active monomer. <ref name="voet" /> The phosphate group on C3 of the substrate is then transferred back onto His 8, thus regenerating the active form of the enzyme. |
| - | In addition to the importance of the two histidine residues in the active site, the amino acids that line the <scene name=' | + | In addition to the importance of the two histidine residues in the active site, the amino acids that line the <scene name='40/401494/Cv/4'>active site</scene> are also functionally important. These residues include H179, H8, E15, S11, T20, R59, and E86.<ref name="voet" /> Several positively charged residues line the active site pocket. These residues usually tend to be <scene name='40/401494/Cv/5'>arginine residues</scene>, which are important for the optimal activity of the enzyme. <ref name="winn" /> This structure is logical for its function because the enzyme binds a negatively charged substrate, thus a positively charged groove fosters tight binding with a negative substrate. The third and final important aspect of the active site is the presence of <scene name='40/401494/Cv/6'>glutamate residues 15 and 86</scene>.<ref name="winn" /> It is suggested that the carboxyl groups of these amino acid residues act as proton-withdrawing groups as they flank both sides of the substrate. |
== Kinetics == | == Kinetics == | ||
| Line 33: | Line 33: | ||
==3D structures of phosphoglycerate mutase== | ==3D structures of phosphoglycerate mutase== | ||
| + | [[Phosphoglycerate mutase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | *Phosphoglycerate mutase | ||
| - | |||
| - | **[[3o0t]], [[3mxo]] – hPGM5 residues 90-289 – human<br /> | ||
| - | **[[1yfk]], [[1yjx]], [[4gpi]], [[4gpz]] – hPGM1<br /> | ||
| - | **[[3ezn]] – BpPGM – ''Burkholderia pseudomallei''<br /> | ||
| - | **[[3d8h]] – PGM – ''Cryptosporidium parvum''<br /> | ||
| - | **[[1v7q]], [[1v37]] – PGM – ''Thermus thermophilus''<br /> | ||
| - | **[[1xq9]] - PfPGM – ''Plasmodium falciparum''<br /> | ||
| - | **[[3kkk]] - PfPGM (mutant) <br /> | ||
| - | **[[1rii]] – PGM – ''Mycobacteriumtuberculosis''<br /> | ||
| - | **[[1e58]] – EcPGM – ''Escherichia coli''<br /> | ||
| - | **[[1fzt]] – PGM – Fission yeast – NMR<br /> | ||
| - | **[[5pgm]], [[4pgm]], [[3pgm]] – yPGM – yeast<br /> | ||
| - | |||
| - | *Phosphoglycerate mutase binary complexes | ||
| - | |||
| - | **[[3lnt]] – BpPGM + malonic acid <br /> | ||
| - | **[[3gw8]] – BpPGM + glycerol + VO4<BR /> | ||
| - | **[[3gp5]] - BpPGM + PGA + VO4<BR /> | ||
| - | **[[3fdz]] - BpPGM + PGA + di-PGA<br /> | ||
| - | **[[3gp3]] – BpPGM + phosphoserine<br /> | ||
| - | **[[1eqj]], [[1ejj]] – BsPGM + PGA – ''Bacillus stearothermophilus''<br /> | ||
| - | **[[1e59]] – EcPGM + VO4<BR /> | ||
| - | **[[1qhf]] – yPGM + PGA <br /> | ||
| - | **[[1bq3]] – yPGM + inositol hexakisphosphate<br /> | ||
| - | **[[1bq4]] – yPGM + benzene hexacarboxylate | ||
| - | |||
| - | *2,3-bisphosphoglycerate-independent phosphoglycerate mutase | ||
| - | |||
| - | **[[1t8p]], [[3nfy]] – hPGM <br /> | ||
| - | **[[4emb]] – PGM – ''Borrelia burgdorferi''<br /> | ||
| - | **[[4eo9]] – PGM – ''Mycobacterium leprae''<br /> | ||
| - | **[[4my4]] – SaPGM – ''Staphylococcus aureus''<br /> | ||
| - | **[[4nwj]] – SaPGM + 3PG<br /> | ||
| - | **[[4nwx]], [[4qax]] – SaPGM + 2PG<br /> | ||
| - | **[[2a9j]], [[2h4x]], [[2h52]] – hPGM + 3PG<br /> | ||
| - | **[[2f90]] - hPGM + 3PG + AlF4<br /> | ||
| - | **[[2h4z]], [[2hhj]] – hPGM + 2,3-BGP<br /> | ||
| - | **[[1ejj]] - GsPGM + 3PG – ''Geobacillus stearothermophilus''<br /> | ||
| - | **[[1eqj]] - GsPGM + 2PG<br /> | ||
| - | **[[3idd]], [[3kd8]] – TaBIPGM – ''Thermoplasma acidophilum''<br /> | ||
| - | **[[3nvl]] – BIPGM - ''Trypanosoma brucei''<br /> | ||
| - | **[[2ify]] – BIPGM – ''Bacillus anthracis''<br /> | ||
| - | **[[2zkt]] – BIPGM – ''Pyrococcus horikoshii''<br /> | ||
| - | **[[3igy]], [[3igz]] – BIPGM + Co + PGA – ''Leishmania mexicana''<br /> | ||
| - | **[[1o98]] – BsBIPGM + PGA<br /> | ||
| - | **[[1o99]] - BsBIPGM (mutant) + PGA | ||
| - | }} | ||
==Additional Resources== | ==Additional Resources== | ||
| - | For additional information, see: [[Carbohydrate Metabolism]] | + | For additional information, please see: [[Carbohydrate Metabolism]] |
<br /> | <br /> | ||
Current revision
| |||||||||||
Additional Resources
For additional information, please see: Carbohydrate Metabolism
References
- ↑ Crowhurst GS, Dalby AR, Isupov MN, Campbell JW, Littlechild JA. Structure of a phosphoglycerate mutase:3-phosphoglyceric acid complex at 1.7 A. Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1822-6. PMID:10531478
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 S., Winn I., Fothergill A. L., Harkins N. R., and Watson C. H. "Structure and Activity of Phosphoglycerate Mutase." Sciences 293.1063 (1981): 121-30. Print.
- ↑ "Phosphoglycerate mutase -." Wikipedia, the free encyclopedia. Web. 27 Feb. 2010. <http://en.wikipedia.org/wiki/Phosphoglycerate_mutase>.
- ↑ 5.0 5.1 5.2 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. Print.
- ↑ Rose, Z.B. (1980) Adv. Enzymol. Relat. Areas Mol. Biol. 51, 211-253
- ↑ Rigden, D. J.; Walter, R. A.; Phillips, S. E. V.; Fothergill-Gilmore, L. A.Polyanionic inhibitors of phosphoglycerate mutase: combined structural and biochemical analysis J. Mol. Biol. 1999, 289, 691– 699
- ↑ McAleese, S.M., Fothergill-Gilmore, L.A.&Dixon, H.B.F. (1985) Biochem. J. 230, 535-542
- ↑ http://www.mda.org/disease/pgam.html
- ↑ http://disability.ucdavis.edu/disease_deatails.php?id=45
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Robert Trahin, Xuan Loi, David Canner, Christopher Vachon, Allie Paton