|
|
| (5 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| | + | |
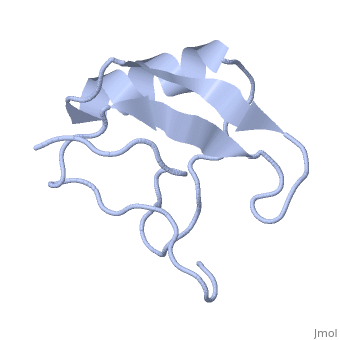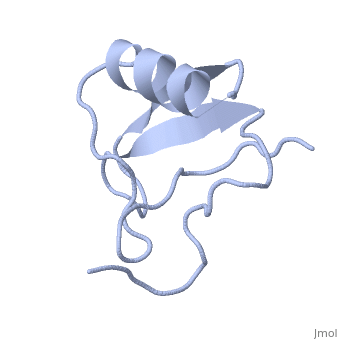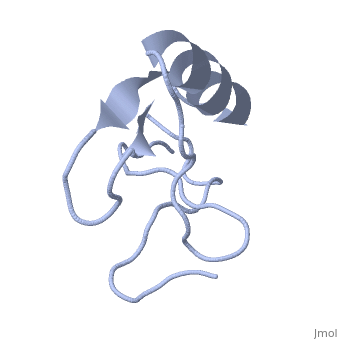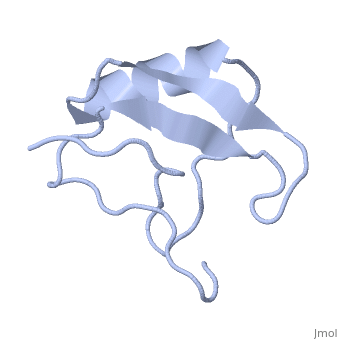
| | ==INSECTICIDAL ALPHA SCORPION TOXIN ISOLATED FROM THE VENOM OF SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, MINIMIZED AVERAGE STRUCTURE== | | ==INSECTICIDAL ALPHA SCORPION TOXIN ISOLATED FROM THE VENOM OF SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, MINIMIZED AVERAGE STRUCTURE== |
| - | <StructureSection load='1lqh' size='340' side='right' caption='[[1lqh]], [[NMR_Ensembles_of_Models | 1 NMR models]]' scene=''> | + | <StructureSection load='1lqh' size='340' side='right'caption='[[1lqh]]' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[1lqh]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Leiqh Leiqh]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1LQH OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1LQH FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[1lqh]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Leiurus_hebraeus Leiurus hebraeus]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1LQH OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1LQH FirstGlance]. <br> |
| - | </td></tr><tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1lqi|1lqi]]</td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR, 1 model</td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1lqh FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1lqh OCA], [http://pdbe.org/1lqh PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1lqh RCSB], [http://www.ebi.ac.uk/pdbsum/1lqh PDBsum]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1lqh FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1lqh OCA], [https://pdbe.org/1lqh PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1lqh RCSB], [https://www.ebi.ac.uk/pdbsum/1lqh PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1lqh ProSAT]</span></td></tr> |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[http://www.uniprot.org/uniprot/SCXA_LEIQH SCXA_LEIQH]] Alpha toxins bind voltage-independently at site-3 of sodium channels (Nav) and inhibit the inactivation of the activated channels, thereby blocking neuronal transmission. The dissociation is voltage-dependent. This toxin is active on insects. It is also highly toxic to crustaceans and has a measurable but low toxicity to mice. | + | [https://www.uniprot.org/uniprot/SCXA_LEIHE SCXA_LEIHE] Alpha toxins bind voltage-independently at site-3 of sodium channels (Nav) and inhibit the inactivation of the activated channels, thereby blocking neuronal transmission. The dissociation is voltage-dependent. This toxin is active on insects. It is also highly toxic to crustaceans and has a measurable but low toxicity to mice. |
| | == Evolutionary Conservation == | | == Evolutionary Conservation == |
| | [[Image:Consurf_key_small.gif|200px|right]] | | [[Image:Consurf_key_small.gif|200px|right]] |
| | Check<jmol> | | Check<jmol> |
| | <jmolCheckbox> | | <jmolCheckbox> |
| - | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/lq/1lqh_consurf.spt"</scriptWhenChecked> | + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/lq/1lqh_consurf.spt"</scriptWhenChecked> |
| - | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> |
| | <text>to colour the structure by Evolutionary Conservation</text> | | <text>to colour the structure by Evolutionary Conservation</text> |
| | </jmolCheckbox> | | </jmolCheckbox> |
| Line 29: |
Line 30: |
| | | | |
| | ==See Also== | | ==See Also== |
| | + | *[[Hemolysin 3D structures|Hemolysin 3D structures]] |
| | *[[Potassium channel toxin|Potassium channel toxin]] | | *[[Potassium channel toxin|Potassium channel toxin]] |
| | + | *[[Potassium channel toxin 3D structures|Potassium channel toxin 3D structures]] |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Leiqh]] | + | [[Category: Large Structures]] |
| - | [[Category: Anglister, J]] | + | [[Category: Leiurus hebraeus]] |
| - | [[Category: Gurevitz, M]] | + | [[Category: Anglister J]] |
| - | [[Category: Kustanovich, I]] | + | [[Category: Gurevitz M]] |
| - | [[Category: Tugarinov, V]] | + | [[Category: Kustanovich I]] |
| - | [[Category: Zilberberg, N]] | + | [[Category: Tugarinov V]] |
| - | [[Category: Neurotoxin]]
| + | [[Category: Zilberberg N]] |
| - | [[Category: Sodium channel inhibitor]]
| + | |
| Structural highlights
Function
SCXA_LEIHE Alpha toxins bind voltage-independently at site-3 of sodium channels (Nav) and inhibit the inactivation of the activated channels, thereby blocking neuronal transmission. The dissociation is voltage-dependent. This toxin is active on insects. It is also highly toxic to crustaceans and has a measurable but low toxicity to mice.
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
The solution structure of a recombinant active alpha-neurotoxin from Leiurus quinquestriatus hebraeus, Lqh(alpha)IT, was determined by proton two-dimensional nuclear magnetic resonance spectroscopy (2D NMR). This toxin is the most insecticidal among scorpion alpha-neurotoxins and, therefore, serves as a model for clarifying the structural basis for their biological activity and selective toxicity. A set of 29 structures was generated without constraint violations exceeding 0.4 A. These structures had root mean square deviations of 0.49 and 1.00 A with respect to the average structure for backbone atoms and all heavy atoms, respectively. Similarly to other scorpion toxins, the structure of Lqh(alpha)IT consists of an alpha-helix, a three-strand antiparallel beta-sheet, three type I tight turns, a five-residue turn, and a hydrophobic patch that includes tyrosine and tryptophan rings in a "herringbone" arrangement. Positive phi angles were found for Ala50 and Asn11, suggesting their proximity to functionally important regions of the molecule. The sample exhibited conformational heterogeneity over a wide range of experimental conditions, and two conformations were observed for the majority of protein residues. The ratio between these conformations was temperature-dependent, and the rate of their interconversions was estimated to be on the order of 1-5 s(-1) at 308 K. The conformation of the polypeptide backbone of Lqh(alpha)IT is very similar to that of the most active antimammalian scorpion alpha-toxin, AaHII, from Androctonus australis Hector (60% amino acid sequence homology). Yet, several important differences were observed at the 5-residue turn comprising residues Lys8-Cys12, the C-terminal segment, and the mutual disposition of these two regions. 2D NMR studies of the R64H mutant, which is 3 times more toxic than the unmodified Lqh(alpha)IT, demonstrated the importance of the spatial orientation of the last residue side chain for toxicity of Lqh(alpha)IT.
Solution structures of a highly insecticidal recombinant scorpion alpha-toxin and a mutant with increased activity.,Tugarinov V, Kustanovich I, Zilberberg N, Gurevitz M, Anglister J Biochemistry. 1997 Mar 4;36(9):2414-24. PMID:9054546[1]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Tugarinov V, Kustanovich I, Zilberberg N, Gurevitz M, Anglister J. Solution structures of a highly insecticidal recombinant scorpion alpha-toxin and a mutant with increased activity. Biochemistry. 1997 Mar 4;36(9):2414-24. PMID:9054546 doi:10.1021/bi961497l
|