We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) Polymerase
From Proteopedia
(Difference between revisions)
| (17 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load='' size='350' side='right' caption='Structure of Yeast Poly(A) Polymerase with ATP, Mg+2 ion (green) and oligo(A) (PDB entry [[2q66]])' scene='Proteopedia:Main_page_develop/2q66_initial/3'> | ||
[[Image:2q66.png|left|170px]] | [[Image:2q66.png|left|170px]] | ||
| + | |||
| + | __TOC__ | ||
==Function== | ==Function== | ||
[[Poly(A) Polymerase]] (polynucleotide adenylytransferase, [[EC Number|EC]] 2.7.7.19) is the enzyme responsible for adding a polyadenine tail to the 3' end of a nascent pre-mRNA transcript. Its substrates are ATP and RNA. The poly(A) tail that poly-A polymerase adds to the 3' end of the pre-mRNA transcript is important for nuclear export, translation and stability of the mRNA. | [[Poly(A) Polymerase]] (polynucleotide adenylytransferase, [[EC Number|EC]] 2.7.7.19) is the enzyme responsible for adding a polyadenine tail to the 3' end of a nascent pre-mRNA transcript. Its substrates are ATP and RNA. The poly(A) tail that poly-A polymerase adds to the 3' end of the pre-mRNA transcript is important for nuclear export, translation and stability of the mRNA. | ||
| + | *'''Star-PAP''' selects mRNA targets for polyadenylation<ref>PMID:34576144</ref>. | ||
==Determinants of ATP Recognition== | ==Determinants of ATP Recognition== | ||
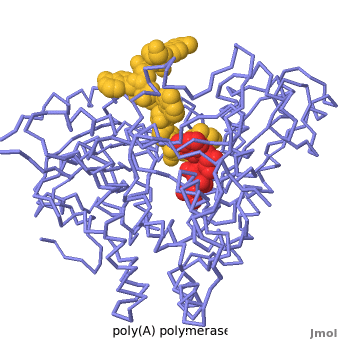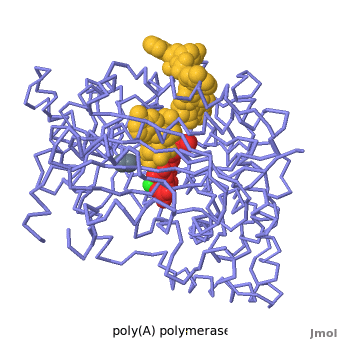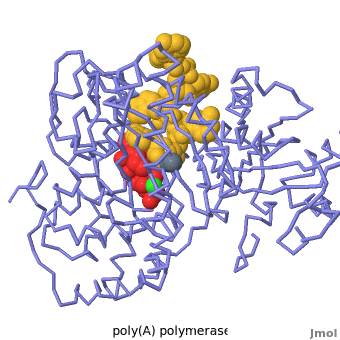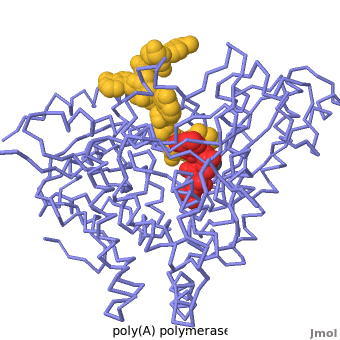
| - | + | Poly(A) Polymerase binds specifically to ATP and adds it at the end of a messenger RNA chain. This structure contains an oligo(A) polynucleotide with five nucleotides, an ATP molecule, and a magnesium ion. The enzyme is an inactive mutant with the catalytic aspartate 154 changed to alanine. Poly(A) polymerase normally has a second magnesium ion, but that second magnesium ion is absent from this structure due to the inactivating mutation D154A. In the <scene name='2q66/2q66_summary/1'>summary picture</scene>, the enzyme is in blue backbone representation, the RNA chain is in yellow, the ATP is in red, the magnesium is in green, and ALA154 is in magenta. Several mechanisms are used to achieve the specificity for ATP. The magnesium is coordinated by <scene name='2q66/2q66_asp/2'>ASP100 and ASP102</scene>, and the magnesium coordinates with the phosphates of ATP, positioning the nucleotide in the active site. The adenine base is sandwiched between the <scene name='2q66/2q66_stacking/2'>terminal base of the RNA (in yellow) and VAL234 (in cyan)</scene>. Surprisingly, there are very few contacts with the hydrogen-bonding groups in the adenine base. <scene name='2q66/2q66_asn/1'>ASN 226</scene> may form a [[hydrogen bond]] to adenine in the active enzyme, but the distance is a bit too long in this mutant structure. Instead of forming specific hydrogen bonds with the enzyme, most of the hydrogen-bonding groups in the base, sugar and phosphate interact with a shell of <scene name='Goodsell_Sandbox/2q66_water/2'>buried water molecules</scene>. Discrimination between ATP and GTP is achieved through a close steric contact between the <scene name='2q66/2q66_c2/2'>adenine C2 (in white) and the sidechains of THR 304 and MET310 (shown in cyan)</scene>. Guanine bases have an extra amino group at this position that would be too bulky to fit against these amino acids. | |
| - | Poly(A) Polymerase binds specifically to ATP and adds it at the end of a messenger RNA chain. This structure contains an oligo(A) polynucleotide with five nucleotides, an ATP molecule, and a magnesium ion. The enzyme is an inactive mutant with the catalytic aspartate 154 changed to alanine. Poly(A) polymerase normally has a second magnesium ion, but that second magnesium ion is absent from this structure due to the inactivating mutation D154A. In the <scene name='2q66/2q66_summary/1'>summary picture</scene>, the enzyme is in blue backbone representation, the RNA chain is in yellow, the ATP is in red, the magnesium is in green, and ALA154 is in magenta. Several mechanisms are used to achieve the specificity for ATP. The magnesium is coordinated by <scene name='2q66/2q66_asp/2'>ASP100 and ASP102</scene>, and the magnesium coordinates with the phosphates of ATP, positioning the nucleotide in the active site. The adenine base is sandwiched between the <scene name='2q66/2q66_stacking/2'>terminal base of the RNA (in yellow) and VAL234 (in cyan)</scene>. Surprisingly, there are very few contacts with the hydrogen-bonding groups in the adenine base. <scene name='2q66/2q66_asn/1'>ASN 226</scene> may form a [[hydrogen bond]] to adenine in the active enzyme, but the distance is a bit too long in this mutant structure. Instead of forming specific hydrogen bonds with the enzyme, most of the hydrogen-bonding groups in the base, sugar and phosphate interact with a shell of <scene name='Goodsell_Sandbox/2q66_water/2'>buried water molecules</scene>. Discrimination between ATP and GTP is achieved through a close steric contact between the <scene name='2q66/2q66_c2/2'>adenine C2 (in white) and the sidechains of THR 304 and MET310 (shown in cyan)</scene>. Guanine bases have an extra amino group at this position that would be too bulky to fit against these amino acids | + | |
| - | + | ||
<blockquote> | <blockquote> | ||
| Line 16: | Line 19: | ||
==3D structures of poly(A) polymerase== | ==3D structures of poly(A) polymerase== | ||
| - | + | [[Poly(A) polymerase 3D structures]] | |
| - | + | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Note: the regulatory subunit of Vaccinia virus Poly(A) polymerase is called '''VP39''' and the catalytic subunit name is '''VP55''' | ||
==Additional Resources== | ==Additional Resources== | ||
| Line 62: | Line 33: | ||
==Reference== | ==Reference== | ||
Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis., Balbo PB, Bohm A, Structure. 2007 Sep;15(9):1117-31. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17850751 17850751] | Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis., Balbo PB, Bohm A, Structure. 2007 Sep;15(9):1117-31. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/17850751 17850751] | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
[[Category: Polynucleotide adenylyltransferase]] | [[Category: Polynucleotide adenylyltransferase]] | ||
Current revision
| |||||||||||
Note: the regulatory subunit of Vaccinia virus Poly(A) polymerase is called VP39 and the catalytic subunit name is VP55
Additional Resources
For additional information, see: Translation
For additional examples of transferases, see: Transferase
- PDB entry 2q66
Reference
Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis., Balbo PB, Bohm A, Structure. 2007 Sep;15(9):1117-31. PMID:17850751
- ↑ Koshre GR, Shaji F, Mohanan NK, Mohan N, Ali J, Laishram RS. Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association. Int J Mol Sci. 2021 Sep 15;22(18):9980. PMID:34576144 doi:10.3390/ijms22189980
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Eran Hodis, David Canner, Joel L. Sussman, OCA, Alexander Berchansky, Jaime Prilusky, David S. Goodsell, Eric Martz