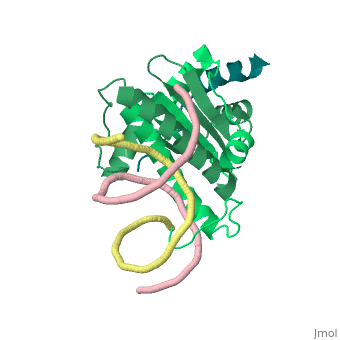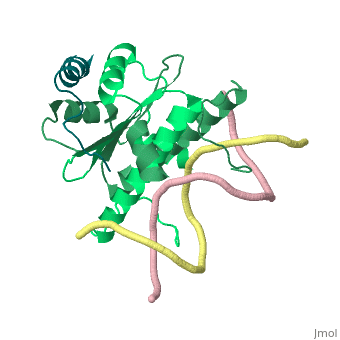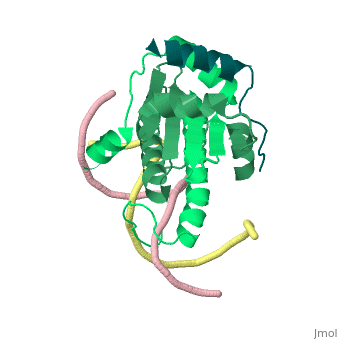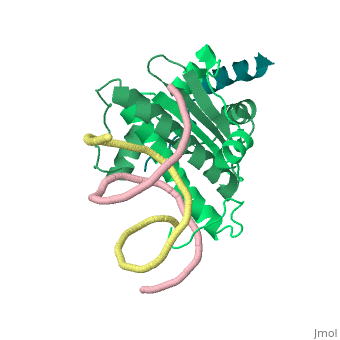Myocyte enhancer factor 2
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | + | <StructureSection load='1tqe' size='350' side='right' caption='Mouse HDAC9 (aquamarine) complex with myocyte-specific enhancer factor (green) and DNA (PDB entry [[1tqa]])' scene=''> | |
| - | =Understanding of the Recruitment of HDACs by MEF2, Based on Their Structure= | + | ==Understanding of the Recruitment of HDACs by MEF2, Based on Their Structure== |
| - | A biological field that has recently gained status is the so called Epigenetics. This term refers to changes in phenotype (appearance) or gene expression caused by mechanisms other than changes in the underlying DNA sequence. Among many factors that act as epigenetic regulators, we can find some proteins that deal with histone ornamentation, such as the antagonists histone acetyltranferases (HATs)[http://en.wikipedia.org/wiki/Histone_acetyltransferase] and the '''histone deacetylases''' (HDACs)[http://en.wikipedia.org/wiki/HDAC]. These proteins act | + | A biological field that has recently gained status is the so called Epigenetics. This term refers to changes in phenotype (appearance) or gene expression caused by mechanisms other than changes in the underlying DNA sequence. Among many factors that act as epigenetic regulators, we can find some proteins that deal with histone ornamentation, such as the antagonists histone acetyltranferases (HATs)[http://en.wikipedia.org/wiki/Histone_acetyltransferase] and the '''histone deacetylases''' (HDACs)[http://en.wikipedia.org/wiki/HDAC]. These proteins act respectively inserting/removing acetyl groups to lysine residues that are contained in histone proteins, making chromatin structures tighter/looser and as a result, promote/repress gene expression<ref>PMID:9798649</ref>. |
Among the subgroups of HDACs, we can cite the class IIa HDACs. Such proteins shuttle from nucleus to cytoplasm in a Ca++ dependent fashion and need to associate to other transcriptional factors, such as MEF2, to have their nuclear activity, because they are unable to anchor to the chromosomes by themselves. | Among the subgroups of HDACs, we can cite the class IIa HDACs. Such proteins shuttle from nucleus to cytoplasm in a Ca++ dependent fashion and need to associate to other transcriptional factors, such as MEF2, to have their nuclear activity, because they are unable to anchor to the chromosomes by themselves. | ||
| Line 14: | Line 13: | ||
Additionally, by looking to the <scene name='Sandbox_Reserved_1/False_dimer/2'>structure</scene> obtained by X-ray crystallography, one could also think that complexes HDAC9/MEF2/DNA dimerize ''in vivo''. During the crystallization process, two complexes were contained in the same asymmetric unit, giving such a false impression. Thus, it is important to emphasize that the biological unit would be composed only by <scene name='Sandbox_Reserved_1/Complex/5'>a monomer of HDAC9, a dimer of MEF2 and a fragment of double strand DNA</scene>[http://www.ebi.ac.uk/pdbe/pqs/pqs-bin/macmol.pl?filename=1TQE]. | Additionally, by looking to the <scene name='Sandbox_Reserved_1/False_dimer/2'>structure</scene> obtained by X-ray crystallography, one could also think that complexes HDAC9/MEF2/DNA dimerize ''in vivo''. During the crystallization process, two complexes were contained in the same asymmetric unit, giving such a false impression. Thus, it is important to emphasize that the biological unit would be composed only by <scene name='Sandbox_Reserved_1/Complex/5'>a monomer of HDAC9, a dimer of MEF2 and a fragment of double strand DNA</scene>[http://www.ebi.ac.uk/pdbe/pqs/pqs-bin/macmol.pl?filename=1TQE]. | ||
| + | |||
| + | *'''MEF2A''' is a substrate for p38<ref>PMID:9858528</ref>. | ||
| + | *'''MEF2B''' and '''MEF2D''' are potent trans-activators expressed in early myogenic lineages<ref>PMID:8669199</ref>. | ||
| + | *'''MEF2C''' controls chondrocyte hypertrophy and bone development<ref>PMID:17336904</ref>. | ||
| + | |||
| + | == Disease == | ||
| + | High expression of MEF2C is observed in leukemia<ref>PMID:23435431</ref>. Mutations in MEF2A are associated with coronary artery disease and myocardial infraction<ref>PMID:20031581</ref>. Mutations in MEF2B are associated with non-hodgkin lymphoma<ref>PMID:26245647</ref>. | ||
==Additional Resources== | ==Additional Resources== | ||
For Additional information, See [[Transcription and RNA Processing]] | For Additional information, See [[Transcription and RNA Processing]] | ||
| + | </StructureSection> | ||
| + | == 3D Structures of myocyte enhancer factor 2 == | ||
| + | |||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[1egw]], [[1c7u]], [[3kov]] – hMEF2A N terminal + DNA – human<br /> | ||
| + | [[3mu6]] – hMEF2A N terminal (mutant) + DNA <br /> | ||
| + | [[3p57]] – hMEF2A N terminal + DNA + histone acetyltransferase<br /> | ||
| + | [[6c9l]] – hMEF2B <br /> | ||
| + | [[1n6j]] – hMEF2B N terminal + DNA + calcineurin-binding protein<br /> | ||
| + | [[1tqe]] – hMEF2B N terminal + DNA + histone deacylase 9 peptide<br /> | ||
| + | [[6wc5]] – hMEF2B + NKX-2.5 + DNA<br /> | ||
| + | [[7x1n]] – hMEF2D + DNA<br /> | ||
| + | [[5f28]] – MEF2C N terminal + focal adhesion kinase 1 - mouse<br /> | ||
| + | == References == | ||
| + | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D Structures of myocyte enhancer factor 2
Updated on 10-July-2023
1egw, 1c7u, 3kov – hMEF2A N terminal + DNA – human
3mu6 – hMEF2A N terminal (mutant) + DNA
3p57 – hMEF2A N terminal + DNA + histone acetyltransferase
6c9l – hMEF2B
1n6j – hMEF2B N terminal + DNA + calcineurin-binding protein
1tqe – hMEF2B N terminal + DNA + histone deacylase 9 peptide
6wc5 – hMEF2B + NKX-2.5 + DNA
7x1n – hMEF2D + DNA
5f28 – MEF2C N terminal + focal adhesion kinase 1 - mouse
References
- ↑ Swanson BJ, Jack HM, Lyons GE. Characterization of myocyte enhancer factor 2 (MEF2) expression in B and T cells: MEF2C is a B cell-restricted transcription factor in lymphocytes. Mol Immunol. 1998 Jun;35(8):445-58. PMID:9798649
- ↑ Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, Olson EN, Ulevitch RJ, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999 Jan;19(1):21-30. PMID:9858528
- ↑ Hornstein A, Batts KP, Linz LJ, Chang CD, Galvanek EG, Bardawil RG. Fine needle aspiration diagnosis of ciliated hepatic foregut cysts: a report of three cases. Acta Cytol. 1996 May-Jun;40(3):576-80. PMID:8669199
- ↑ Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007 Mar;12(3):377-89. PMID:17336904 doi:http://dx.doi.org/10.1016/j.devcel.2007.02.004
- ↑ Cante-Barrett K, Pieters R, Meijerink JP. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014 Jan 23;33(4):403-10. doi: 10.1038/onc.2013.56. Epub 2013 Feb 25. PMID:23435431 doi:http://dx.doi.org/10.1038/onc.2013.56
- ↑ Guella I, Rimoldi V, Asselta R, Ardissino D, Francolini M, Martinelli N, Girelli D, Peyvandi F, Tubaro M, Merlini PA, Mannucci PM, Duga S. Association and functional analyses of MEF2A as a susceptibility gene for premature myocardial infarction and coronary artery disease. Circ Cardiovasc Genet. 2009 Apr;2(2):165-72. doi:, 10.1161/CIRCGENETICS.108.819326. Epub 2009 Feb 12. PMID:20031581 doi:http://dx.doi.org/10.1161/CIRCGENETICS.108.819326
- ↑ Pon JR, Wong J, Saberi S, Alder O, Moksa M, Grace Cheng SW, Morin GB, Hoodless PA, Hirst M, Marra MA. MEF2B mutations in non-Hodgkin lymphoma dysregulate cell migration by decreasing MEF2B target gene activation. Nat Commun. 2015 Aug 6;6:7953. doi: 10.1038/ncomms8953. PMID:26245647 doi:http://dx.doi.org/10.1038/ncomms8953