Acetylcholinesterase
From Proteopedia
(Difference between revisions)
| (93 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' scene='22/22/Ache_with_ach/2' caption='Torpedo california AChE (PDB code [[2ace]])'> |
[[Image:small_wh_ray0001.gif|left|150px]]<br /> | [[Image:small_wh_ray0001.gif|left|150px]]<br /> | ||
'''Acetylcholinesterase''' (AChE) is key enzyme in the nervous system of animals. By rapid hydrolysis of the neurotransmitter, [[Acetylcholine|acetylcholine]] (ACh), AChE terminates neurotransmission at cholinergic synapses. It is a very fast enzyme, especially for a serine hydrolase, functioning at a rate approaching that of a diffusion-controlled reaction. AChE inhibitors are among the key drugs approved by the FDA for management of Alzheimer's disease (AD). The powerful toxicity of organophosphorus (OP) poisons is attributed primarily to their potent AChE inhibitors. | '''Acetylcholinesterase''' (AChE) is key enzyme in the nervous system of animals. By rapid hydrolysis of the neurotransmitter, [[Acetylcholine|acetylcholine]] (ACh), AChE terminates neurotransmission at cholinergic synapses. It is a very fast enzyme, especially for a serine hydrolase, functioning at a rate approaching that of a diffusion-controlled reaction. AChE inhibitors are among the key drugs approved by the FDA for management of Alzheimer's disease (AD). The powerful toxicity of organophosphorus (OP) poisons is attributed primarily to their potent AChE inhibitors. | ||
| + | |||
| + | See also [[Acetylcholinesterase (Hebrew)]] | ||
== Key Enzyme in the Nervous System == | == Key Enzyme in the Nervous System == | ||
Solution of the three-dimensional (3D) structure of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] (''Tc''AChE) in 1991 opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation.<ref>PMID:1678899</ref> The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis.<ref>PMID:10545346</ref> To understand how those aromatic residues behave with the enzyme, see [[Flexibility of aromatic residues in acetylcholinesterase]]. Solution of the 3D structure of acetylcholinesterase led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | Solution of the three-dimensional (3D) structure of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] (''Tc''AChE) in 1991 opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation.<ref>PMID:1678899</ref> The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis.<ref>PMID:10545346</ref> To understand how those aromatic residues behave with the enzyme, see [[Flexibility of aromatic residues in acetylcholinesterase]]. Solution of the 3D structure of acetylcholinesterase led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | ||
| - | [http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | + | [http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression systems. |
| - | systems. | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | {{Clear}} | ||
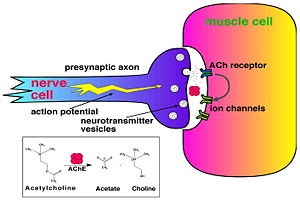
[[Image:Synapse_Schematic.jpg|thumb|Cholinergic Synapse|300px|left]] | [[Image:Synapse_Schematic.jpg|thumb|Cholinergic Synapse|300px|left]] | ||
| - | + | ||
| + | [http://en.wikipedia.org/wiki/Acetylcholinesterase Acetylcholinesterase] [http://en.wikipedia.org/wiki/Hydrolysis hydrolysizes] the [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] [http://en.wikipedia.org/wiki/Acetylcholine acetylcholine] <scene name='2ace/Cv/2'>(ACh)</scene>, producing <scene name='2ace/Cv/3'>choline and an acetate</scene> group. ACh directly binds <scene name='22/22/Cv/1'>Ser200</scene> (via its [http://en.wikipedia.org/wiki/Nucleophile nucleophilic] Oγ atom) within the <scene name='2ace/Cv/5'>catalytic triad (Ser200, His440, and Glu327)</scene> (ACh/''Tc''AChE structure [[2ace]]). The residues <scene name='2ace/Cv/6'>Trp84 and Phe330</scene> are also important in the [http://en.wikipedia.org/wiki/Ligand ligand] recognition <ref name="Raves">PMID:8989325</ref>. After this binding acetylcholinesterase <scene name='2ace/Cv/7'>hydrolysizes</scene> ACh. <br /> | ||
| + | See also [[Acetylcholinesterase with acetylcholine]]. | ||
| + | |||
== Treatment of Alzheimer's disease == | == Treatment of Alzheimer's disease == | ||
| - | [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease] (AD) is a disorder that attacks the [http://en.wikipedia.org/wiki/Central_nervous_system central nervous system] through progressive degeneration of its neurons. AD occurs in around 10% of the elderly and, as yet, there is no known cure. Patients with this disease develop [http://en.wikipedia.org/wiki/Dementia dementia] which becomes more severe as the disease progresses. It was suggested that symptoms of AD are caused by decrease of activity of [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Neocortex neocortical] and [http://en.wikipedia.org/wiki/Hippocampus hippocampal] neurons. Treatment of AD by ACh precursors and [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Agonist agonists] was ineffective or caused severe side effects. ACh hydrolysis by AChE causes termination of cholinergic neurotransmission. Therefore, compounds which inhibit AChE might significantly increase the levels of ACh depleted in AD. Indeed, it was shown that [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitors] improve the cognitive abilities of AD patients at early stages of the disease development. | + | [http://en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease] (AD) is a disorder that attacks the [http://en.wikipedia.org/wiki/Central_nervous_system central nervous system] through progressive degeneration of its neurons. AD occurs in around 10% of the elderly and, as yet, there is no known cure. Patients with this disease develop [http://en.wikipedia.org/wiki/Dementia dementia] which becomes more severe as the disease progresses. It was suggested that symptoms of AD are caused by decrease of activity of [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Neocortex neocortical] and [http://en.wikipedia.org/wiki/Hippocampus hippocampal] neurons. Treatment of AD by ACh precursors and [http://en.wikipedia.org/wiki/Cholinergic cholinergic] [http://en.wikipedia.org/wiki/Agonist agonists] was ineffective or caused severe side effects. ACh hydrolysis by AChE causes termination of cholinergic neurotransmission. Therefore, compounds which inhibit AChE might significantly increase the levels of ACh depleted in AD. Indeed, it was shown that [http://en.wikipedia.org/wiki/Acetylcholinesterase_inhibitor AChE inhibitors] improve the cognitive abilities of AD patients at early stages of the disease development. The way in which the various cholinesterase inhibitors interact with AChE can be see at:<br /> |
| - | + | *[[Acetylcholinesterase: Treatment of Alzheimer's disease]].<br /> | |
| - | + | *[[Acetylcholinesterase complexed with N-9-(1',2',3',4'-tetrahydroacridinyl)-1,8-diaminooctane]].<br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Organophosphorus acid anhydride nerve agents == | == Organophosphorus acid anhydride nerve agents == | ||
| Line 73: | Line 26: | ||
At the <scene name='2wfz/Ali/3'>active site of the nonaged soman/TcAChE conjugate</scene> ([[2wfz]]) the catalytic His440 forms hydrogen bonds with Ser200 Oγ and Glu327 Oε1 via its Nε2 and Nδ1 nitrogens, respectively. The O2 atom of soman is within hydrogen bonding distance of His440 Nε2. Soman O1 mimicks carbonyl oxygen of ACh. A water molecule 1001 interacting with soman O2 is represented as a <font color='red'><b>red ball</b></font>. The active site residues of the nonaged soman/TcAChE are colored <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. The O2 atom of the <scene name='2wfz/Ali/4'>dealkylated (aged) soman</scene> ([[2wg0]]) forms a salt bridge with His440 Nε2. The active site residues of the aged soman/TcAChE are colored <span style="color:pink;background-color:black;font-weight:bold;">pink</span>. <scene name='2wfz/Ali/5'>Alignment</scene> of the structures of the nonaged ([[2wfz]]) and aged ([[2wg0]]) conjugates reveals a small, but important, change within the active site - the imidazole ring of His440 is tilted back to a native-like conformation after dealkylation. The water molecule 1001, which interacts with soman O2 in the nonaged crystal structure, is not within hydrogen bonding distance of O2 in the aged crystal structure. 2-PAM binds poorly to the nonaged phosphonylated enzyme (its electron density was not found) and binds in an <scene name='2wfz/Ali/7'>unfavorable and nonfunctional conformation</scene> after soman aging to ''Tc''AChE ([[2wg1]]) <ref name="Sanson">PMID:19642642</ref>. | At the <scene name='2wfz/Ali/3'>active site of the nonaged soman/TcAChE conjugate</scene> ([[2wfz]]) the catalytic His440 forms hydrogen bonds with Ser200 Oγ and Glu327 Oε1 via its Nε2 and Nδ1 nitrogens, respectively. The O2 atom of soman is within hydrogen bonding distance of His440 Nε2. Soman O1 mimicks carbonyl oxygen of ACh. A water molecule 1001 interacting with soman O2 is represented as a <font color='red'><b>red ball</b></font>. The active site residues of the nonaged soman/TcAChE are colored <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. The O2 atom of the <scene name='2wfz/Ali/4'>dealkylated (aged) soman</scene> ([[2wg0]]) forms a salt bridge with His440 Nε2. The active site residues of the aged soman/TcAChE are colored <span style="color:pink;background-color:black;font-weight:bold;">pink</span>. <scene name='2wfz/Ali/5'>Alignment</scene> of the structures of the nonaged ([[2wfz]]) and aged ([[2wg0]]) conjugates reveals a small, but important, change within the active site - the imidazole ring of His440 is tilted back to a native-like conformation after dealkylation. The water molecule 1001, which interacts with soman O2 in the nonaged crystal structure, is not within hydrogen bonding distance of O2 in the aged crystal structure. 2-PAM binds poorly to the nonaged phosphonylated enzyme (its electron density was not found) and binds in an <scene name='2wfz/Ali/7'>unfavorable and nonfunctional conformation</scene> after soman aging to ''Tc''AChE ([[2wg1]]) <ref name="Sanson">PMID:19642642</ref>. | ||
| - | == | + | ==Additional resources== |
| + | see: [[Acetylcholinesterase_Additional_Resources]] | ||
| - | + | ==Movies== | |
| + | see: [[Acetylcholinesterase_Movies]] | ||
| - | + | ==Acetylcholinesterase 3D structures== | |
| - | + | ||
| - | + | ||
| - | + | [[Acetylcholinesterase 3D structures]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==References== | |
| + | <references/> | ||
</StructureSection> | </StructureSection> | ||
| - | |||
| - | == 3D Structures of AChE == | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | *Acetylcholinesterase - AChE native | ||
| - | |||
| - | **[[3lii]], [[4ey4]] – hAChE - recombinant human <br /> | ||
| - | **[[1ea5]], [[2ace]] – ''Tc''AChE – trigonal – ''Torpedo californica'' <br /> | ||
| - | **[[2j3d]] – ''Tc''AChE – monoclinic <br /> | ||
| - | **[[1w75]] – ''Tc''AChE – orthorhombic <br /> | ||
| - | **[[2vt6]], [[2vt7]] – ''Tc''AChE – different dosage <br /> | ||
| - | **[[1qid]] to [[1qim]] - ''Tc''AChE synchrotron radiation damage <br /> | ||
| - | **[[1j06]], [[1maa]] – mAChE - mouse <br /> | ||
| - | **[[1qo9]] – ''Dm''AChE - ''Drosophila melanogaster'' <br /> | ||
| - | **[[1eea]], [[1c2b]], [[1c2o]] – AChE – Electric eel <br /> | ||
| - | |||
| - | *AChE inhibitors (In Different Languages) | ||
| - | |||
| - | **[[1eve]] AChE-Aricept complex, [[1eve (Arabic)]], [[1eve (Chinese)]], [[1eve (Italian)]], [[1eve (Russian)]], [[1eve (Spanish)]], [[1eve (Turkish)]], [[1eve (Italian)]] <br /> | ||
| - | **[[1vot]] AChE-Huperzine A complex, [[1vot (Chinese)]] <br /> | ||
| - | |||
| - | *AChE active site inhibitors conjugating at the bottom of the active site gorge | ||
| - | |||
| - | **[[2c4h]] – ''Tc''AChE + acetylthiocholine<br /> | ||
| - | **[[2w9i]] – ''Tc''AChE + methylene blue <br /> | ||
| - | **[[2wls]] – MosAChE + AMTS13 <br /> | ||
| - | **[[2vq6]] – ''Tc''AChE + 2-PAM <br /> | ||
| - | **[[2j3q]] – ''Tc''AChE + Thioflavin T <br /> | ||
| - | **[[2ha0]] – mAChE + ketoamyltrimethylammonium <br /> | ||
| - | **[[2h9y]] – mAChE + TMTFA <br /> | ||
| - | **[[3zlt]] – mAChE + RVX<br /> | ||
| - | **[[3zlu]] – mAChE + cyclosarin<br /> | ||
| - | **[[3zlv]] – mAChE + tabun + HI-6<br /> | ||
| - | **[[4bc0]], [[4bc1]] – mAChE + CBDP<br /> | ||
| - | **[[1gpk]], [[1gpn]], [[1vot]] – ''Tc''AChE + huperzine <br /> | ||
| - | **[[4ey5]] – hAChE + huperzine<br /> | ||
| - | **[[1gqr]] – ''Tc''AChE + rivastigmine <br /> | ||
| - | **[[1gqs]] – ''Tc''AChE + NAP <br /> | ||
| - | **[[1e66]] – ''Tc''AChE + huprine <br /> | ||
| - | **[[4a16]] – mAChE + huprine <br /> | ||
| - | **[[1dx4]], [[1qon]] – ''Dm''AChE + tacrine derivative <br /> | ||
| - | **[[1oce]] – ''Tc''AChE + MF268 <br /> | ||
| - | **[[1ax9]], [[1ack]] – ''Tc''AChE + edrophonium <br /> | ||
| - | **[[1amn]] – ''Tc''AChE + TMTFA <br /> | ||
| - | **[[1acj]] – ''Tc''AChE + tacrine <br /> | ||
| - | **[[4tvk]] - TcAChE + tacrine hybrid<br /> | ||
| - | **[[1u65]] – ''Tc''AChE + CPT-11<br /> | ||
| - | **[[2bag]] - ''Tc''AChE + ganstigmine<br /> | ||
| - | **[[2xi4]] - ''Tc''AChE + aflatoxin<br /> | ||
| - | **[[4ara]], [[4arb]], [[4a23]], [[4b7z]], [[4b80]], [[4b81]], [[4b82]], [[4b83]], [[4b84]], [[4b85]], [[4btl]] - mAChE + inhibitor<br /> | ||
| - | **[[2xuf]], [[2xug]], [[2xuh]], [[2xui]], [[2xuj]], [[2xuk]], [[2xuo]], [[2xup]], [[2xuq]] - mAChE (mutant) + inhibitor<br /> | ||
| - | **[[4m0e]], [[4m0f]] - hAChE + inhibitor<br /> | ||
| - | |||
| - | *AChE peripheral site inhibitors conjugating at the surface of the protein | ||
| - | |||
| - | **[[1ku6]], [[1mah]] - mAChE + fasciculin 2 <br /> | ||
| - | **[[1j07]] - mAChE + decidium <br /> | ||
| - | **[[1n5m]] - mAChE + gallamine <br /> | ||
| - | **[[1n5r]] - mAChE + propidium <br /> | ||
| - | **[[1b41]], [[1f8u]], [[4ey8]] - hAChE + fasciculin 2 <br /> | ||
| - | **[[1fss]] - TcAChE + fasciculin 2 <br /> | ||
| - | **[[2x8b]] - hAChE + fasciculin 2 + tabun<br /> | ||
| - | **[[4bdt]] - hAChE + fasciculin 2 + huprine W<br /> | ||
| - | |||
| - | *AChE bis inhibitors spanning the active site gorge | ||
| - | |||
| - | **[[3i6m]] – ''Tc''AChE + N-piperidinopropyl galanthamine <br /> | ||
| - | **[[3i6z]] - ''Tc''AChE + saccharinohexyl galanthamine <br /> | ||
| - | **[[1zgb]], [[1zgc]] – ''Tc''AChE + tacrine (10) hupyridone <br /> | ||
| - | **[[2w6c]] – ''Tc''AChE + bis-(-)-nor-meptazinol <br /> | ||
| - | **[[2ckm]], [[2cmf]] – ''Tc''AChE + bis-tacrine <br /> | ||
| - | **[[2cek]] – ''Tc''AChE + N-[8-(1,2,3,4-tetrahydroacridin-9-ylthio)octyl]-1,2,3,4-tetrahydroacridin-9-amine <br /> | ||
| - | **[[1ut6]] - ''Tc''AChE + N-9-(1,2,3,4-tetrahydroacridinyl)-1,8-diaminooctane <br /> | ||
| - | **[[1odc]] - ''Tc''AChE + N-4-quinolyl-N-9-(1,2,3,4-tetrahydroacridinyl)-1,8-diaminooctane <br /> | ||
| - | **[[1w4l]], [[1w6r]], [[1w76]], [[1dx6]], [[1qti]] - TcAChE + galanthamine and derivative <br /> | ||
| - | **[[4ey6]] - hAChE + galanthamine<br /> | ||
| - | **[[1q83]], [[1q84]] - mAChE + TZ2PA6 <br /> | ||
| - | **[[1h22]], [[1h23]] – ''Tc''AChE + bis-hupyridone <br /> | ||
| - | **[[1hbj]] – ''Tc''AChE + quinoline derivativev <br /> | ||
| - | **[[1e3q]] – ''Tc''AChE + bw284c51 <br /> | ||
| - | **[[1eve]] – ''Tc''AChE + e2020 <br /> | ||
| - | **[[1acl]] – ''Tc''AChE + decamethonium <br /> | ||
| - | **[[2xud]] – TcAChE (mutant) + decamethonium<br /> | ||
| - | **[[3zv7]] - ''Tc''AChE + bisnorcymserine | ||
| - | |||
| - | *AChE organophosphate inhibitors causing irreversible inhibition | ||
| - | |||
| - | **[[2wu3]] – mAChE + fenamiphos and HI-6 <br /> | ||
| - | **[[2wu4]] – mAChE + fenamiphos and ortho-7 <br /> | ||
| - | **[[2jgf]] - mAChE + fenamiphos <br /> | ||
| - | **[[2wfz]], [[2wg0]], [[2wg2]], [[1som]] - ''Tc''AChE + soman <br /> | ||
| - | **[[2wg1]] - ''Tc''AChE + soman + 2-PAM <br /> | ||
| - | **[[2whp]], [[2whq]], [[2whr]] – mAChE + sarin and HI-6 <br /> | ||
| - | **[[2jgg]], [[2y2v]] - mAChE + sarin <br /> | ||
| - | **[[2jgl]] - mAChE + VX and sarin <br /> | ||
| - | **[[1cfj]] - ''Tc''AChE + sarin, GB <br /> | ||
| - | **[[3dl4]], [[3dl7]] – mAChE + tabun <br /> | ||
| - | **[[2jey]] – mAChE + HLO-7 <br /> | ||
| - | **[[2c0p]], [[2c0q]] - mAChE + tabun <br /> | ||
| - | **[[2jez]] - mAChE + tabun + HLO-7 <br /> | ||
| - | **[[2jf0]] - mAChE + tabun + Ortho-7 <br /> | ||
| - | **[[2jgh]], [[2y2u]] - mAChE + VX <br /> | ||
| - | **[[1vxo]], [[1vxr]] - ''Tc''AChE + VX<br /> | ||
| - | **[[2jgi]], [[2jgm]] - mAChE + DFP <br /> | ||
| - | **[[1dfp]] - ''Tc''AChE + DFP <br /> | ||
| - | **[[2jgj]], [[2jgk]], [[2jge]] - mAChE + methamidophos <br /> | ||
| - | **[[2gyu]] - mAChE + HI-6 <br /> | ||
| - | **[[2gyv]] - mAChE + Ortho-7 <br /> | ||
| - | **[[2gyw]] - mAChE + obidoxime <br /> | ||
| - | **[[3gel]] - TcAChE + methyl paraoxon<br /> | ||
| - | **[[2dfp]] – TcAChE aged<br /> | ||
| - | |||
| - | *AChE substrate analogues mimicking the binding of the substrate acetylcholine | ||
| - | |||
| - | **[[2ha4]] – mAChE (mutant) + acetylcholine <br /> | ||
| - | **[[2vja]], [[2vjb]], [[2vjc]], [[2vjd]], [[2cf5]] – TcAChE + 4-oxo-N,N,N-trimethylpentanaminium | ||
| - | **[[2v96]], [[2v97]], [[2v98]], [[2v99]] – ''Tc''AChE + 1-(2-nitrophenyl)-2,2,2-trifluoroethyl-arsenocholine <br /> | ||
| - | **[[2ha2]] – mAChE + succinylcholine <br /> | ||
| - | **[[2ha3]] - mAChE + choline <br /> | ||
| - | **[[2ha5]] – mAChE (mutant) + acetylthiocholine <br /> | ||
| - | **[[2ha6]] – mAChE (mutant) + succinylthiocholine <br /> | ||
| - | **[[2ha7]] – mAChE (mutant) + butyrylthiocholine <br /> | ||
| - | **[[2ch4]], [[2c58]] – ''Tc''AChE + acetylthiocholine <br /> | ||
| - | **[[2c5g]] – ''Tc''AChE + thiocholine <br /> | ||
| - | **[[2c5f]] – TcAChE + substrate analog<br /> | ||
| - | **[[2va9]] - TcAChE + ‘caged’ arsenocholine<br /> | ||
| - | **[[4ey7]] – hAChE + donepezil<br /> | ||
| - | |||
| - | *Others... | ||
| - | |||
| - | **[[2j4f]] – ''Tc''AChE + Hg <br /> | ||
| - | **[[1vzj]] – ''Tc''AChE tetramerization domain <br /> | ||
| - | **[[1jjb]] – ''Tc''AChE + PEG <br /> | ||
| - | **[[1qie]], [[1qif]], [[1qig]], [[1qih]], [[1qii]], [[1qij]], [[1qik]] – TcAChE synchrotron radiation damage <br /> | ||
| - | **[[3m3d]] – TcAChE + Xe<br /> | ||
| - | **[[4qww]] – AChE + antibody – banded krait<br /> | ||
| - | }} | ||
| - | |||
| - | ==Additional Resources== | ||
| - | For additional information, see:<br /> | ||
| - | [[Alzheimer's Disease]]<br /> | ||
| - | [[AChE inhibitors and substrates]]<br /> | ||
| - | |||
| - | |||
[[Category: catalytic triad]] | [[Category: catalytic triad]] | ||
[[Category: cholinesterase]] | [[Category: cholinesterase]] | ||
| - | [[Category: cholinesterases]] | ||
[[Category: acetylcholine]] | [[Category: acetylcholine]] | ||
[[Category: cation-pi]] | [[Category: cation-pi]] | ||
| Line 266: | Line 48: | ||
[[Category: nerve gasses]] | [[Category: nerve gasses]] | ||
| - | ==External Links == | ||
| - | *[http://www.messiah.edu/departments/chemistry/molscilab/jtat_080120/acetylcholinesterase/contents/contents.htm Acetylcholinesterase Tutorial] by Karl Oberholser, Messiah College | ||
| - | *[http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb54_1.html PDB Molecule of the Month - Acetylcholinesterase] | ||
| - | *[http://www.weizmann.ac.il/sb/faculty_pages/Sussman/movies/Radiation_Damage Movies: X-ray Damage in ACh] & [http://www.weizmann.ac.il/sb/faculty_pages/Sussman/movies/richardnew.mpg Nature's Vacuum Cleaner] by R. Gillilan, Cornell Univ | ||
| - | ==References== | ||
| - | |||
| - | <references/> | ||
| - | [[Treatments:AChE Inhibitor References]]<br /> | ||
| - | [[Treatments:Alzheimer's Disease]] | ||
| - | [[he:Acetylcholinesterase_(Hebrew)]] | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Alexander Berchansky, David Canner, Eran Hodis, Clifford Felder, Jaime Prilusky, Harry Greenblatt, Yechun Xu