We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1160
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 9: | Line 9: | ||
The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | ||
| - | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The structure of the alpha helices are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator mavoglurant shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 -<scene name='72/721531/Protien_lys/3'>Lysozyme</scene> was inserted into intracellular loop 2 to add stability<ref name="Dore" />. | + | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The structure of the <font color='darkgreen'>'''alpha helices'''</font> are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator <span style="color:yellow;background-color:black;font-weight:bold;">mavoglurant</span> shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 -<scene name='72/721531/Protien_lys/3'>Lysozyme</scene> was inserted into intracellular loop 2 to add stability<ref name="Dore" />. |
== Structure== | == Structure== | ||
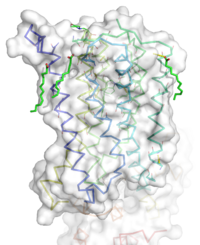
[[Image:STR.png|200 px|left|thumb|'''Figure 1''': Overall Structure of the mGlu5 TMD. The polar heads on the Oleic acids orient the image with the top oriented extracellularly, the middle portion inserted into the membrane, and the lower portion oriented intracellularly. The white exterior represents the surface of the protien, and the multicolored lines interior to the surface represent the backbones 7 transmembrane alpha helices. ]] | [[Image:STR.png|200 px|left|thumb|'''Figure 1''': Overall Structure of the mGlu5 TMD. The polar heads on the Oleic acids orient the image with the top oriented extracellularly, the middle portion inserted into the membrane, and the lower portion oriented intracellularly. The white exterior represents the surface of the protien, and the multicolored lines interior to the surface represent the backbones 7 transmembrane alpha helices. ]] | ||
| Line 20: | Line 20: | ||
=== Binding Pocket === | === Binding Pocket === | ||
[[Image: Organic with clipped surface.png|200 px|left|thumb|'''Figure 2.''' Mavoglurant in its binding pocket of the 7TM region of mGLu5 Class C receptor. The binding pocket's surface is clipped in black with the substrate, mavoglurant, in red. The rest of the protein is colored in green. The binding pocket is present in the 7 Helix Transmembrane Domain that would be present in the phospholipid bilayer as an integral protein. The presence of mavoglurant inhibits the function of the metabotropic glutamate receptor.]] | [[Image: Organic with clipped surface.png|200 px|left|thumb|'''Figure 2.''' Mavoglurant in its binding pocket of the 7TM region of mGLu5 Class C receptor. The binding pocket's surface is clipped in black with the substrate, mavoglurant, in red. The rest of the protein is colored in green. The binding pocket is present in the 7 Helix Transmembrane Domain that would be present in the phospholipid bilayer as an integral protein. The presence of mavoglurant inhibits the function of the metabotropic glutamate receptor.]] | ||
| - | The binding pocket, not to be confused with the glutamate binding site, represents a useful source of regulatory control. The binding pocket is only accessible by a relatively narrow (7 Å) <scene name='72/721531/Bindingsite/ | + | The binding pocket, not to be confused with the glutamate binding site, represents a useful source of regulatory control. The binding pocket is only accessible by a relatively narrow (7 Å) <scene name='72/721531/Bindingsite/2'>entrance</scene><ref name="Dore" />. This small entrance severely restricts the access of both positive and negative allosteric regulators. One such regulator, <scene name='72/721531/Mavo/2'>mavoglurant</scene>, is a relatively small molecule that can travel through the entrance and fit between the helices of the mGlu5 receptor<ref name="Dore" />. Mavoglurant acts as a negative allosteric modulator of the mGlu5 receptor<ref name="Dore" />. Mavoglurant is a relatively hydrophobic molecule, which compliments the largely hydrophobic nature of the interior of mGlu5<ref name="Dore" />. The carbamate tail and hydroxyl group of mavoglurant will also interact with several key amino acids in the binding site of mGlu5<ref name="Dore" />. |
Important Amino Acids<ref name="Dore" />: | Important Amino Acids<ref name="Dore" />: | ||
| - | *<scene name='72/721531/Protien_bindtop/ | + | *<scene name='72/721531/Protien_bindtop/8'>Asparagine</scene> 747 forms a hydrogen bond network with the main chain carbonyl of Glycine 652 and the carbamate portion of mavoglurant. |
*Bicyclic ring of mavoglurant surrounded by <scene name='72/721531/Protien_hydrophobic/1'>hydrophobic binding pocket</scene>. <font color='purple'>'''Hydrophobic regions'''</font> of the mGlu5 receptor are shown in <font color='purple'>'''purple'''</font> and the <font color='grey'>'''rest of the helices'''</font> are shown in <font color='grey'>'''grey'''</font>. Mavoglurant is represented by spheres. | *Bicyclic ring of mavoglurant surrounded by <scene name='72/721531/Protien_hydrophobic/1'>hydrophobic binding pocket</scene>. <font color='purple'>'''Hydrophobic regions'''</font> of the mGlu5 receptor are shown in <font color='purple'>'''purple'''</font> and the <font color='grey'>'''rest of the helices'''</font> are shown in <font color='grey'>'''grey'''</font>. Mavoglurant is represented by spheres. | ||
*2 Catalytic <scene name='72/721531/Protien_bindmiddle/4'>serine</scene> residues H-bond to the hydroxyl oxygen of our ligand. | *2 Catalytic <scene name='72/721531/Protien_bindmiddle/4'>serine</scene> residues H-bond to the hydroxyl oxygen of our ligand. | ||
Current revision
Human metabotropic glutamate receptor 5 transmembrane domain
| |||||||||||