Sandbox Reserved 1180
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 15: | Line 15: | ||
===Structure=== | ===Structure=== | ||
| - | The class B GPCRs, including GCGR, are different from other GPCRs in several ways. The first is that class B GPCRs contain a protrusion known as a 'stalk,' a three α-helical turn elongation of the N-terminus that protrudes past the extracellular (EC) membrane.<ref name= "Siu 2013"/> Structural integrity of this domain in GCGR is <scene name='72/721552/The_right_one/3'>essential to ligand binding affinity.</scene> A135P mutations impact stalk stability by removing an important salt bridge between Glu133 - Lys136.<ref name= "Siu 2013"/> A second difference between class B and other GPCRs is that the extracellular loop 1 (ECL1) is 3-4 times longer than comparable loops in class A GPCRs, and also affects ligand binding affinity.<ref name= "Siu 2013"/> Most notably, class B GPCRs contain a <scene name='72/727091/Corticotropin_glucagon_aligned/3'>prominent central splay</scene> which is solvent filled and accessible from the extracellular side.<ref name= "Hollenstein 2014"/> This central splay is notably absent <scene name='72/721551/B2-adrenergic_glucagon_aligned/2'>from other GPCRs</scene>, and represents a tantalizing target for agonists/antagonists.<ref name= "Hollenstein 2014"/> | + | The class B GPCRs, including [http://www.rcsb.org/pdb/explore/explore.do?structureId=4L6R GCGR], are different from other GPCRs in several ways. The first is that class B GPCRs contain a protrusion known as a 'stalk,' a three α-helical turn elongation of the N-terminus that protrudes past the extracellular (EC) membrane.<ref name= "Siu 2013"/> Structural integrity of this domain in GCGR is <scene name='72/721552/The_right_one/3'>essential to ligand binding affinity.</scene> A135P mutations impact stalk stability by removing an important salt bridge between Glu133 - Lys136.<ref name= "Siu 2013"/> A second difference between class B and other GPCRs is that the extracellular loop 1 (ECL1) is 3-4 times longer than comparable loops in class A GPCRs, and also affects ligand binding affinity.<ref name= "Siu 2013"/> Most notably, class B GPCRs contain a <scene name='72/727091/Corticotropin_glucagon_aligned/3'>prominent central splay</scene> which is solvent filled and accessible from the extracellular side.<ref name= "Hollenstein 2014"/> This central splay is notably absent <scene name='72/721551/B2-adrenergic_glucagon_aligned/2'> from other GPCRs</scene>, and represents a tantalizing target for agonists/antagonists.<ref name= "Hollenstein 2014"/> |
Because of the difficulty in stabilizing and crystallizing Class B TMDs, very little is known about the conformational changes that transduce cell signals endogenously. GCGR is known to regulate additional signal pathways through the adoption of differing receptor conformations and to interact with receptor activity-modifying proteins (RAMPs) altering the signaling bias of the receptor.<ref name= "Xu 2009"/> | Because of the difficulty in stabilizing and crystallizing Class B TMDs, very little is known about the conformational changes that transduce cell signals endogenously. GCGR is known to regulate additional signal pathways through the adoption of differing receptor conformations and to interact with receptor activity-modifying proteins (RAMPs) altering the signaling bias of the receptor.<ref name= "Xu 2009"/> | ||
| Line 21: | Line 21: | ||
===Glucagon Binding=== | ===Glucagon Binding=== | ||
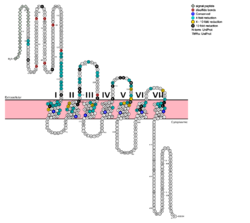
[[Image:Protter GLR HUMAN.png |250 px|left|thumb|Fig. 1: Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.<ref name= "Siu 2013"/>]] | [[Image:Protter GLR HUMAN.png |250 px|left|thumb|Fig. 1: Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.<ref name= "Siu 2013"/>]] | ||
| - | [[Image:Movie Frame 6.png |150 px|right|thumb|Fig. 2: <scene name='72/721552/Glucagon_binding/3'>Deep, central cavity</scene> functioning as anchoring site for glucagon's n-terminal residues.]][[Image:Glucagon with Q3 and N-terminus.png |150 px|right|thumb|Fig. 3: Surface visualization of glucagon visualizing the three dimensional shape of the N-terminal tail that interacts with the binding site of GCGR central cavity.]] The large, soluble N-terminal extracellular domains (ECD) of GCGR provide initial ligand selectivity with the deep, ligand pocket (Fig. 2) of the TMD providing secondary recognition.<ref name= "Yang 2015"/> In a comprehensive mutagenesis and glucagon-binding study, a total of 129 mutations of GCGR were tested. 41 of these covering 28 different locations in the GCGR TMD were found to have at least a fourfold decrease in glucagon binding affinity<ref name= "Siu 2013"/>. (see Fig. 1) It is the face of the central cavity that harbors the majority of the residues which play an important role in glucagon binding.<ref name= "Siu 2013"/> The binding site was shown to be a dynamic area traveling from the middle of the stalk region (Tyr 138) to deep within the 7TM core (Glu 362), encompassing positions along ECL1, ECL2 and ECL3 and helices I, II, III, V, VI and VII. | + | [[Image:Movie Frame 6.png |150 px|right|thumb|Fig. 2: <scene name='72/721552/Glucagon_binding/3'>Deep, central cavity</scene> functioning as anchoring site for glucagon's n-terminal residues.]][[Image:Glucagon with Q3 and N-terminus.png |150 px|right|thumb|Fig. 3: Surface visualization of glucagon visualizing the three dimensional shape of the N-terminal tail that interacts with the binding site of GCGR central cavity.]] The large, soluble N-terminal extracellular domains (ECD) of GCGR provide initial ligand selectivity with the deep, ligand pocket (Fig. 2) of the TMD providing secondary recognition.<ref name= "Yang 2015"/> In a comprehensive mutagenesis and glucagon-binding study, a total of 129 mutations of GCGR were tested. 41 of these covering 28 different locations in the GCGR TMD were found to have at least a fourfold decrease in glucagon binding affinity<ref name= "Siu 2013"/>. (see Fig. 1) It is the face of the central cavity that harbors the majority of the residues which play an important role in glucagon binding.<ref name= "Siu 2013"/> The binding site was shown to be a [http://proteopedia.org/wiki/index.php/Image:Movie_Frame_8.png dynamic area] traveling from the middle of the stalk region (Tyr 138) to deep within the 7TM core (Glu 362), encompassing positions along ECL1, ECL2 and ECL3 and helices I, II, III, V, VI and VII. |
Mutagenesis and photo cross-linking studies determined essential, conserved residues in glucagon and have been <scene name='72/727091/Glucagon_important_residues/2'>labeled and colored</scene> in red.<ref name= "Siu 2013"/> Glucagon residues His 1, Gln 3, Phe 6, and Tyr 10 are critical to successful binding interaction with the GCGR while others are important for structural rigidity. The n-terminus of glucagon (Fig. 3) leads to a protuberance that fits into the deep, interior cavity of the GCGR 7TMD (Fig. 2) where four residues reside that play strong roles in ligand binding affinity. There is a <scene name='72/721552/Glucagon_binding_zoomed_in/1'>narrow neck</scene> to the entrance of the cavity, providing a firm anchor during peptide docking. (also see Fig. 2) | Mutagenesis and photo cross-linking studies determined essential, conserved residues in glucagon and have been <scene name='72/727091/Glucagon_important_residues/2'>labeled and colored</scene> in red.<ref name= "Siu 2013"/> Glucagon residues His 1, Gln 3, Phe 6, and Tyr 10 are critical to successful binding interaction with the GCGR while others are important for structural rigidity. The n-terminus of glucagon (Fig. 3) leads to a protuberance that fits into the deep, interior cavity of the GCGR 7TMD (Fig. 2) where four residues reside that play strong roles in ligand binding affinity. There is a <scene name='72/721552/Glucagon_binding_zoomed_in/1'>narrow neck</scene> to the entrance of the cavity, providing a firm anchor during peptide docking. (also see Fig. 2) | ||
Current revision
Glucagon G protein-coupled receptors
| |||||||||||