Human Erythrocyte Catalase
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='1dgb' size='350' side='right' caption='Human Erythrocyte Catalase (pdb code 1dgb)' scene=''> | <StructureSection load='1dgb' size='350' side='right' caption='Human Erythrocyte Catalase (pdb code 1dgb)' scene=''> | ||
| - | == | + | == Introduction == |
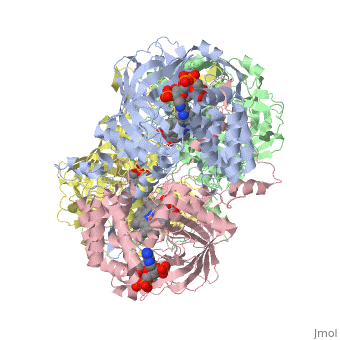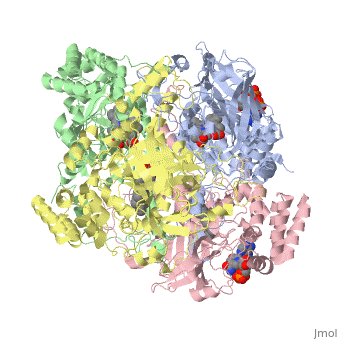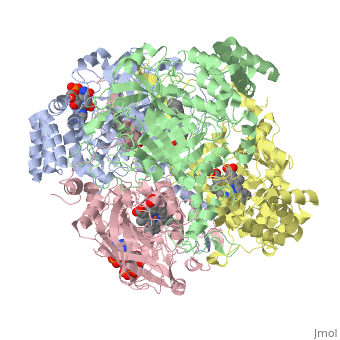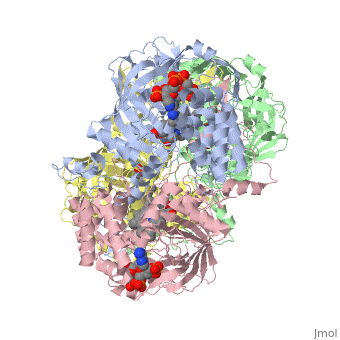
| - | Human Erythrocyte Catalase is an enzyme whose main function is to break down hydrogen peroxide that is produced as a byproduct of metabolism<ref name="yazdi">PMID:25739358</ref>. Catalase can break down approximately one million molecules of hydrogen peroxide per second <ref name="Alfonso-Prietro" />. Without catalase hydrogen peroxide would build up and cause damage to the cell and could even signal it to initiate cell death <ref name="yazdi" />. Structurally it contains four subunits each with a heme group in the center which allows it to break down the peroxide in human cells <ref name="Alfonso-Prietro" />. | + | Human Erythrocyte Catalase is an enzyme whose main function is to break down hydrogen peroxide that is produced as a byproduct of metabolism<ref name="yazdi">PMID:25739358</ref>. Catalase can break down approximately one million molecules of hydrogen peroxide per second <ref name="Alfonso-Prietro" />. Without catalase hydrogen peroxide would build up and cause damage to the cell and could even signal it to initiate cell death <ref name="yazdi" />. Structurally it contains four subunits each with a heme group in the center which allows it to break down the peroxide in human cells <ref name="Alfonso-Prietro" />. See also [[Catalase]]. |
== Function == | == Function == | ||
| Line 18: | Line 18: | ||
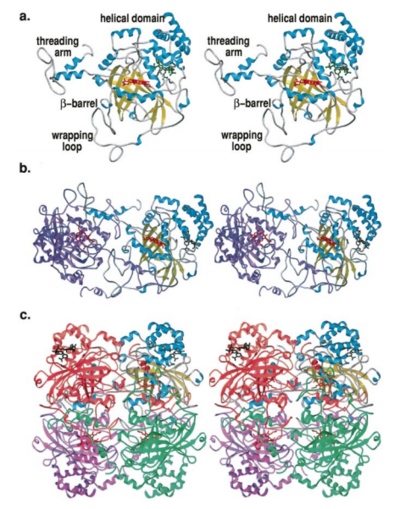
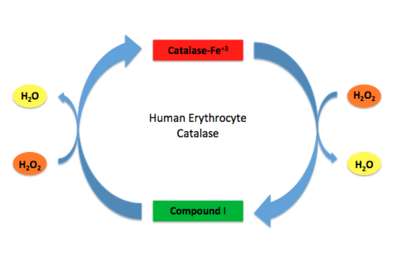
Human erythrocyte catalase is a negatively charged heme-containing monofunctional tetrameric enzyme that is prevalent among aerobic organisms <ref name= "Kodydková" >PMID:25152049</ref><ref name=Alfonso-Prietro>PMID:22516655</ref><ref name=Dash>PMID:22521743</ref><ref name=Diaz>PMID:22209752 </ref><ref name=Nishikawa>PMID:19385054 </ref>. The catalase fold contains an eight-sheeted anti-parallel beta-barrel domain linked to a six alpha-helical domain via a lengthy protein sequence. Residues within β1-β4 contribute to the heme variant, while residues within β5-β8 establish the NADPH binding site <ref name="Diaz" />. The positioning of the heme is determined by the proximal aromatic pyrrole compounds; in human erythrocyte catalase, catalytic His75 is <scene name='72/728053/Position_of_his75/3'>positioned</scene> above pyrrole ring III, further producing a His-III orientation and heme-b variant. The <scene name='72/728053/Active_site_1/2'>NADPH Binding site </scene> is located at the β,α-domain junction <ref name="Alfonso-Prietro" /><ref name="Diaz" />. When the NADPH molecule is bound, a right-handed clockwise helical formation is produced. In human erythrocyte catalase, only two of the four subunits allow for NADPH binding <ref name="Kodydková" /><ref name="Diaz" />. The active site contains a negatively charged tyrosine and a positively charged histidine situated, respectively, proximal and distal to the heme group. The histidine is responsible for the formation of Compound I during the first step of the catalase mechanism <ref name="Alfonso-Prietro" />. | Human erythrocyte catalase is a negatively charged heme-containing monofunctional tetrameric enzyme that is prevalent among aerobic organisms <ref name= "Kodydková" >PMID:25152049</ref><ref name=Alfonso-Prietro>PMID:22516655</ref><ref name=Dash>PMID:22521743</ref><ref name=Diaz>PMID:22209752 </ref><ref name=Nishikawa>PMID:19385054 </ref>. The catalase fold contains an eight-sheeted anti-parallel beta-barrel domain linked to a six alpha-helical domain via a lengthy protein sequence. Residues within β1-β4 contribute to the heme variant, while residues within β5-β8 establish the NADPH binding site <ref name="Diaz" />. The positioning of the heme is determined by the proximal aromatic pyrrole compounds; in human erythrocyte catalase, catalytic His75 is <scene name='72/728053/Position_of_his75/3'>positioned</scene> above pyrrole ring III, further producing a His-III orientation and heme-b variant. The <scene name='72/728053/Active_site_1/2'>NADPH Binding site </scene> is located at the β,α-domain junction <ref name="Alfonso-Prietro" /><ref name="Diaz" />. When the NADPH molecule is bound, a right-handed clockwise helical formation is produced. In human erythrocyte catalase, only two of the four subunits allow for NADPH binding <ref name="Kodydková" /><ref name="Diaz" />. The active site contains a negatively charged tyrosine and a positively charged histidine situated, respectively, proximal and distal to the heme group. The histidine is responsible for the formation of Compound I during the first step of the catalase mechanism <ref name="Alfonso-Prietro" />. | ||
| + | |||
| Line 36: | Line 37: | ||
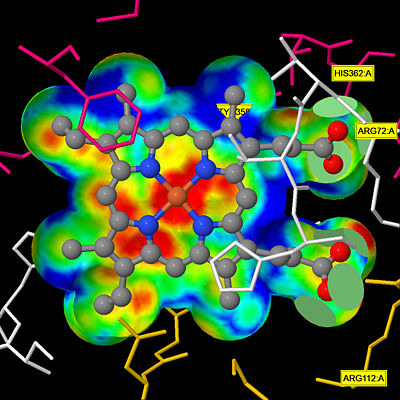
The deeply buried heme group is connected to the protein surface by a primary channel which provides a transport pathway for the hydrogen peroxide substrate <ref name="Diaz" />. The transportation of hydrogen peroxide through the main channel is regulated by electrical dipole interactions between the hydrogen peroxide and the hydrophobic portion of the channel containing negatively charged aspartate and positively charged iron from the heme <ref name="Lennicke" /><ref name="Diaz" />. Additionally, less significant lateral channels allow products to leave the heme pocket<ref name="Diaz" />. | The deeply buried heme group is connected to the protein surface by a primary channel which provides a transport pathway for the hydrogen peroxide substrate <ref name="Diaz" />. The transportation of hydrogen peroxide through the main channel is regulated by electrical dipole interactions between the hydrogen peroxide and the hydrophobic portion of the channel containing negatively charged aspartate and positively charged iron from the heme <ref name="Lennicke" /><ref name="Diaz" />. Additionally, less significant lateral channels allow products to leave the heme pocket<ref name="Diaz" />. | ||
| + | |||
| + | [[Image:HEC jsmol.jpg|thumb|center|400px|'''Heme Pocket Interaction''' This figure illustrates a protein channel that creates a "pocket" from the surface of the molecule to the heme group deep in the molecule that allows hydrogen peroxide molecules to be transported through.<ref>Putnam, C.D., Arvai, A.S., Bourne, Y., Tainer, J.A.(2000). 1DGB HUMAN ERYTHROCYTE CATALASE. [http://www.rcsb.org/pdb/explore/explore.do?structureId=1DGB DOI: 10.2210/pdb1dgb/pdb]</ref> ]] | ||
Current revision
1dgb
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Lindsey Bittle, Brianna Easton, Alyssa Gard, Alexander Berchansky, Rachel Gaunt, Jessica Smyth