We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
CYP3A4
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load='4NY4' size='400' side='right' scene='' caption='Human heme-containing cytochrome P450 3A4 complex with inhibitor (PDB code [[4ny4]])'> | ||
== Overview == | == Overview == | ||
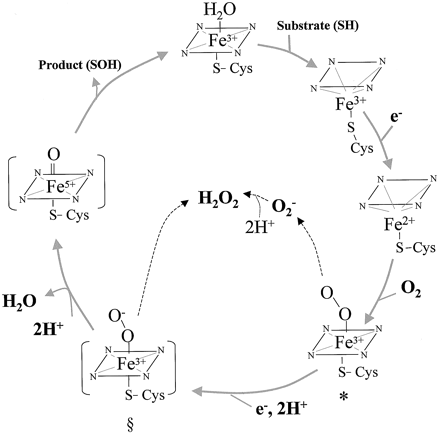
| - | + | '''Cytochrome P450 3A4''' (CYP3A4) is in the heme-thiolate monooxygenase enzyme family meaning the protein contains a heme group with an iron atom. Enzymes in [[Cytochrome P450]] are oxidizing enzymes and CYP3A4 works in the body oxidizing foreign molecules such as toxins and drugs<ref name="a">PMID:16389357</ref><ref name="b">PMID:15603755</ref>. It appears almost half of the marketed pharmaceutical drugs are metabolized by CYP3A4 [http://www.pharmacytimes.com/publications/issue/2008/2008-09/2008-09-8687] All Cytochrome P450 are essential enzymes for metabolism and the enzyme CYP3A4 is the most important. [http://www.medsafe.govt.nz/profs/PUArticles/March2014DrugMetabolismCytochromeP4503A4.htm] Primarily found in the liver and intestine, CYP3A4 is localized in the endoplasmic reticulum membrane [http://www.uniprot.org/uniprot/P08684] and are present in all eukaryotic organisms as well as some prokaryotes<ref name="c">PMID: 15352783</ref>. While CYP3A4's role in drug metabolism are numerous, they are often aiding deactivation through facilitated excretion from the system or by direct inactivation. [https://en.wikipedia.org/wiki/CYP3A4#Tissue_distribution] | |
| - | + | ||
| - | + | ||
| Line 22: | Line 21: | ||
The gene encoding CYP3A4 is located on chromosome 7 in the human genome<ref name="e">PMID: 1391968</ref> and it has been found that there are significant variants of the protein correlating to race <ref name="f"> PMID: 11714865</ref>. This finding is relevant due to the proteins altered ability to react with substrates such as testosterone <ref name="g">PMID: 11714865</ref>. | The gene encoding CYP3A4 is located on chromosome 7 in the human genome<ref name="e">PMID: 1391968</ref> and it has been found that there are significant variants of the protein correlating to race <ref name="f"> PMID: 11714865</ref>. This finding is relevant due to the proteins altered ability to react with substrates such as testosterone <ref name="g">PMID: 11714865</ref>. | ||
| - | The | + | The <scene name='72/728174/N_to_c_terminus/1'>alpha helices</scene> can be seen colored in rainbow succession from the N to C terminus. The protein pocket, where the reactions are catalyzed, more specifically, the <scene name='72/728174/Heme_binding_site/1'>heme binding site</scene> contains twenty-two residues [http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=4NY4] but the main interaction is with a Cysteine at position 442, which coordinates with iron. [http://www.uniprot.org/uniprot/P08684]. |
| + | == Drug Metabolism == | ||
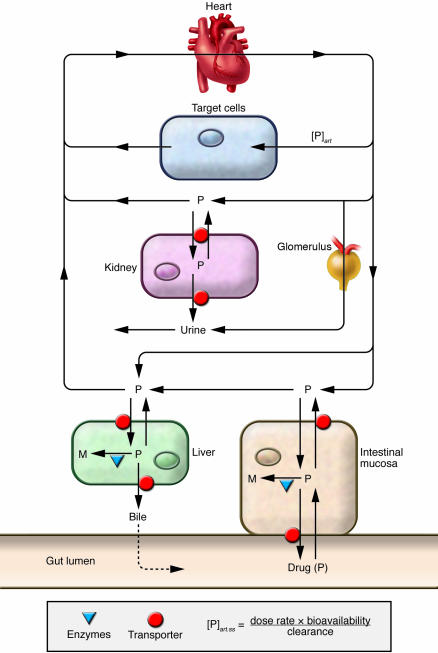
| + | CYP3A4 is thought to affect the metabolism of 30-60% of the drugs on the market [https://www.pharmgkb.org/gene/PA130?tabType=tabVip#tabview=tab3&subtab=32] [http://www.pharmacytimes.com/publications/issue/2008/2008-09/2008-09-8687]. An orally prescribed drug can take a while to arrive at its target cell and along the way the body has enzymes that filter out the drug dampening its efficacy. Of those enzymes, CYP3A4 is a main player and still a lot more needs to be learned. In many cases, like that of the ß2 receptor, there is only a fraction of the drug remaining after what is known as the first-pass metabolism so that by the time the drug gets to the circulatory system where it can be delivered to the tissues its potency has already been compromised <ref name="h"> PMCID: PMC2045626</ref>. | ||
| + | One way to work around this is by intravenous administration so as to bypass the biological filters of the gut and go right into the bloodstream. | ||
| + | [[Image:First-pass metabolism.jpg]] | ||
| + | </StructureSection> | ||
| + | <b>References</b><br> | ||
| + | <references/> | ||
| - | + | [[Category:Topic Page]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| |||||||||||
References
- ↑ Ogu CC, Maxa JL. Drug interactions due to cytochrome P450. Proc (Bayl Univ Med Cent). 2000 Oct;13(4):421-3. PMID:16389357
- ↑ Gonzalez FJ. Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat Res. 2005 Jan 6;569(1-2):101-10. PMID:15603755 doi:http://dx.doi.org/10.1016/j.mrfmmm.2004.04.021
- ↑ Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004 Sep;104(9):3947-80. PMID:15352783 doi:http://dx.doi.org/10.1021/cr020443g
- ↑ Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004 Sep;104(9):3947-80. PMID:15352783 doi:http://dx.doi.org/10.1021/cr020443g
- ↑ Devlin, Thomas M., ed. Textbook of Biochemistry with Clinical Correlations. 6th ed. Hoboken: John Wiley, 2006. Print.
- ↑ Inoue K, Inazawa J, Nakagawa H, Shimada T, Yamazaki H, Guengerich FP, Abe T. Assignment of the human cytochrome P-450 nifedipine oxidase gene (CYP3A4) to chromosome 7 at band q22.1 by fluorescence in situ hybridization. Jpn J Hum Genet. 1992 Jun;37(2):133-8. PMID:1391968 doi:http://dx.doi.org/10.1007/BF01899734
- ↑ Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW, Goldstein JA. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001 Dec;299(3):825-31. PMID:11714865
- ↑ Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW, Goldstein JA. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001 Dec;299(3):825-31. PMID:11714865
- ↑ PMCID: PMC2045626