Methylamine dehydrogenase
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
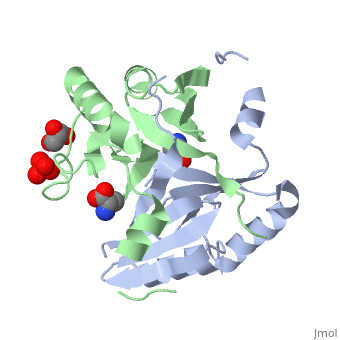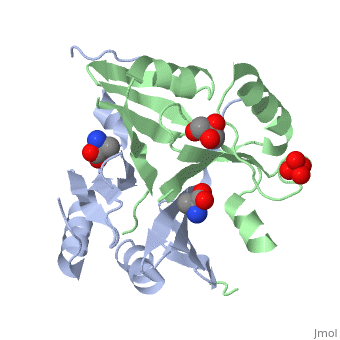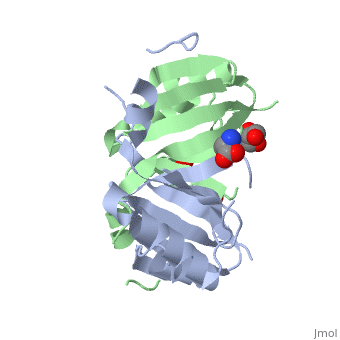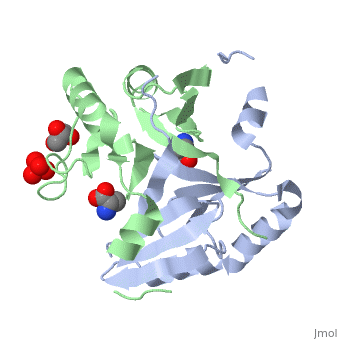
| - | <StructureSection load=' | + | <StructureSection load='4nb0' size='350' side='right' caption='Methylamine dehydrogenase tetramer: 2 heavy chains (salmon and blue) and 2 light chains (green and yellow) complex with tryptophylquinone (TTQ) (PDB entry [[2bbk]])' scene=''> |
== Function == | == Function == | ||
'''Methylamine dehydrogenase''' (MADH) catalyzes the oxidative deamination of primary amine to aldehyde and ammonia, in particular, the conversion of methylamine to formaldehyde. MADH is tryptophan tryptophyl-quinone (TTQ) dependent. MADH is a heterotetramer containing 2 heavy (α) and 2 light (β) subunits. Each β subunit contains a TTQ prosthetic group. The posttranslational modification of two tryptophan residues in '''preMADH''' to form the TTQ cofactor of MADH is catalyzed by methylation utilization protein (MauG). MADH forms a complex with cytochrome c-551i. In the complex, electrons are transferred from TTQ via the amicyanin copper ion center to the heme group of cytochrome<ref>PMID:3558322</ref>. | '''Methylamine dehydrogenase''' (MADH) catalyzes the oxidative deamination of primary amine to aldehyde and ammonia, in particular, the conversion of methylamine to formaldehyde. MADH is tryptophan tryptophyl-quinone (TTQ) dependent. MADH is a heterotetramer containing 2 heavy (α) and 2 light (β) subunits. Each β subunit contains a TTQ prosthetic group. The posttranslational modification of two tryptophan residues in '''preMADH''' to form the TTQ cofactor of MADH is catalyzed by methylation utilization protein (MauG). MADH forms a complex with cytochrome c-551i. In the complex, electrons are transferred from TTQ via the amicyanin copper ion center to the heme group of cytochrome<ref>PMID:3558322</ref>. | ||
== Structural highlights == | == Structural highlights == | ||
| - | The MADH unique redox center <scene name='49/490916/Cv/ | + | The MADH unique redox center <scene name='49/490916/Cv/4'>TTQ cofactor</scene> is located in the light subunit<ref>PMID:9514722</ref>. <scene name='49/490916/Cv/5'>Whole redox center</scene>. |
</StructureSection> | </StructureSection> | ||
| Line 18: | Line 18: | ||
**[[2mta]], [[2gc7]] - PdMADH α + β + amicyanin + cytochrome c551i<br /> | **[[2mta]], [[2gc7]] - PdMADH α + β + amicyanin + cytochrome c551i<br /> | ||
**[[1mg2]], [[1mg3]], [[2gc4]] - PdMADH α (mutant) + β+ amicyanin + cytochrome c551i<br /> | **[[1mg2]], [[1mg3]], [[2gc4]] - PdMADH α (mutant) + β+ amicyanin + cytochrome c551i<br /> | ||
| - | **[[3rn1]], [[3sle]], [[3svw]], [[3sws]], [[3sxt]], [[4k3i]], [[4o1q]] - PdMADH α + β + MauG<br /> | + | **[[3rn1]], [[3sle]], [[3svw]], [[3sws]], [[3sxt]], [[4k3i]], [[4o1q]], [[4y5r]] - PdMADH α + β + MauG<br /> |
**[[3sjl]] - PdMADH α + β (mutant) + MauG<br /> | **[[3sjl]] - PdMADH α + β (mutant) + MauG<br /> | ||
Current revision
| |||||||||||
3D structures of methylamine dehydrogenase
Updated on 11-July-2019
References
- ↑ Husain M, Davidson VL. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J Bacteriol. 1987 Apr;169(4):1712-7. PMID:3558322
- ↑ Chen L, Doi M, Durley RC, Chistoserdov AY, Lidstrom ME, Davidson VL, Mathews FS. Refined crystal structure of methylamine dehydrogenase from Paracoccus denitrificans at 1.75 A resolution. J Mol Biol. 1998 Feb 13;276(1):131-49. PMID:9514722 doi:10.1006/jmbi.1997.1511