User:R. Jeremy Johnson/Glutamate Receptor
From Proteopedia
< User:R. Jeremy Johnson(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 11: | Line 11: | ||
The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | ||
| - | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The structure of the <font color='darkgreen'>'''alpha helices'''</font> are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator <span style="color:yellow;background-color:black;font-weight:bold;">mavoglurant</span> shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 - | + | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The <scene name='72/721531/Protien_lys/3'>structure</scene> of the <font color='darkgreen'>'''alpha helices'''</font> are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator <span style="color:yellow;background-color:black;font-weight:bold;">mavoglurant</span> shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 -Lysozyme was inserted into intracellular loop 2 to add stability<ref name="Dore" />. |
== Structure == | == Structure == | ||
| Line 19: | Line 19: | ||
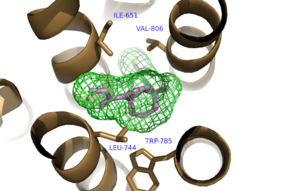
A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>, as demonstrated by <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> being represented in the inactivate state. While in the inactive state, glutamate binding to mGlu<sub>5</sub> triggers a conformational change that leads to mGlu<sub>5</sub> being in the active state and hence initiates the aforementioned [https://en.wikipedia.org/wiki/Gq_alpha_subunit G<sub>q</sub> pathway]. The first of these interactions is an ionic interaction, termed the <scene name='72/726409/Ionic_lock2/2'>Ionic Lock</scene>, between Lysine 665 of TM3 and Glutamate 770 of TM6. Evidence for the importance of this interaction came through a kinetic study of mutant proteins where both residues were separately substituted with alanine, resulting in constitutive activity of the GPCR and its coupled pathway.<ref name="Dore"/> A second critical interaction that stabilizes the inactive conformer is a <scene name='72/726409/Hydrogen_bond_614-668/2'>Hydrogen Bond </scene> between Serine 614 of ICL1 and Arginine 668 of TM3. Similarly, when Serine 614 was substituted with alanine, high levels of activity were seen in the mutant GPCR.<ref name="Dore"/> A <scene name='72/726404/Scene_6/10'>Disulfide Bond </scene> between Cysteine 644 of TM3 and Cysteine 733 of <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene> is critical at anchoring ECL2 and is highly conserved across Class C GPCR’s.<ref name="Dore"/> The ECL2's presence combined with the helical bundle of the trans-membrane domain creates a <scene name='72/726409/Electrogradient2/9'>Binding Cap</scene> that restricts entrance to the allosteric binding site within the seven trans-membrane α-helices. This restricted entrance has no effect on the natural ligand, glutamate, as glutamate binds to the extracellular domain, but this entrance dictates potential drug targets that act through allosteric modulation.<ref name="Dore"/> | A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>, as demonstrated by <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> being represented in the inactivate state. While in the inactive state, glutamate binding to mGlu<sub>5</sub> triggers a conformational change that leads to mGlu<sub>5</sub> being in the active state and hence initiates the aforementioned [https://en.wikipedia.org/wiki/Gq_alpha_subunit G<sub>q</sub> pathway]. The first of these interactions is an ionic interaction, termed the <scene name='72/726409/Ionic_lock2/2'>Ionic Lock</scene>, between Lysine 665 of TM3 and Glutamate 770 of TM6. Evidence for the importance of this interaction came through a kinetic study of mutant proteins where both residues were separately substituted with alanine, resulting in constitutive activity of the GPCR and its coupled pathway.<ref name="Dore"/> A second critical interaction that stabilizes the inactive conformer is a <scene name='72/726409/Hydrogen_bond_614-668/2'>Hydrogen Bond </scene> between Serine 614 of ICL1 and Arginine 668 of TM3. Similarly, when Serine 614 was substituted with alanine, high levels of activity were seen in the mutant GPCR.<ref name="Dore"/> A <scene name='72/726404/Scene_6/10'>Disulfide Bond </scene> between Cysteine 644 of TM3 and Cysteine 733 of <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene> is critical at anchoring ECL2 and is highly conserved across Class C GPCR’s.<ref name="Dore"/> The ECL2's presence combined with the helical bundle of the trans-membrane domain creates a <scene name='72/726409/Electrogradient2/9'>Binding Cap</scene> that restricts entrance to the allosteric binding site within the seven trans-membrane α-helices. This restricted entrance has no effect on the natural ligand, glutamate, as glutamate binds to the extracellular domain, but this entrance dictates potential drug targets that act through allosteric modulation.<ref name="Dore"/> | ||
| - | + | When mGlu5 is in the active conformation, signaling begins with glutamate binding to the Venus flytrap domain. The signal is transduced across the cysteine-rich domain to the TMD.<ref name="Niswender" /> Next, the dimerization of the TMD occurs. This activates the Gq/11 pathway, which activates phospholipase Cβ<ref name="Niswender" />. The active [http://www.proteopedia.org/wiki/index.php/2zkm phospholipase Cβ] hydrolyzes phosphotinositides and generates [https://pubchem.ncbi.nlm.nih.gov/compound/439456#section=Top inositol 1,4,5-trisphosphate] and [http://www.sivabio.50webs.com/ip3.htm diacyl-glycerol].<ref name="Woodcock">PMID: 18940816</ref> This results in calcium mobilization and activation of protein kinase C ([[PKC]])<ref name="Niswender" />. Calcium is a neurotransmitter, and relatively low concentrations of calcium can cause a large response across the neuronal synapse<ref name="Niswender" />. In addition to calcium stimulating an excitory response in nerve cells, PKC can be activated for regulatory purposes by the influx of calcium. A serine on PKC can become phosphorylated, which leaves PKC unable to bind to G beta-gamma (Gßγ) protein complex<ref name="Niswender" />. Unbound Gßγ protein can then inhibit voltage-sensitive calcium channels to reduce calcium influx and provide feedback inhibition to the glutamate signaling pathway. <ref name="Niswender" />. | |
=== Extracellular Domain === | === Extracellular Domain === | ||
| Line 56: | Line 56: | ||
== External Resources == | == External Resources == | ||
| + | |||
| + | [https://www.autismparentingmagazine.com/oppositional-defiant-disorder-treatment/ OOD (Oppositional Defiant Disorder) Treatment Plan] | ||
| + | |||
[http://www.fraxa.org/novartis-discontinues-development-mavoglurant-afq056-fragile-x-syndrome/ Novartis Fragile X trials] | [http://www.fraxa.org/novartis-discontinues-development-mavoglurant-afq056-fragile-x-syndrome/ Novartis Fragile X trials] | ||
| Line 65: | Line 68: | ||
[http://www.en.wikipedia.org/wiki/Gq_alpha_subunit Gq Alpha subunit] | [http://www.en.wikipedia.org/wiki/Gq_alpha_subunit Gq Alpha subunit] | ||
| + | |||
| + | ==Proteopedia Resources== | ||
| + | [http://proteopedia.org/wiki/index.php/Glutamate_receptor Glutamate receptors] | ||
| + | |||
| + | [http://proteopedia.org/wiki/index.php/Metabotropic_glutamate_receptor Metabotropic glutamate receptor] | ||
| + | |||
| + | [http://proteopedia.org/wiki/index.php/Ionotropic_Glutamate_Receptor Ionotropic glutamate receptor] | ||
| + | |||
| + | [http://proteopedia.org/wiki/index.php/User:R._Jeremy_Johnson/CH462:Biochemistry_II_Butler_University Butler University Proteopedia Pages] | ||
| + | </StructureSection> | ||
| + | |||
| + | ==Student Contributors== | ||
| + | Connor Coatney | ||
| + | |||
| + | Kurt Corsbie | ||
| + | |||
| + | Cutter Koehler | ||
| + | |||
| + | Dan Schemenauer | ||
Current revision
| |||||||||||
Student Contributors
Connor Coatney
Kurt Corsbie
Cutter Koehler
Dan Schemenauer