|
|
| (3 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| | | | |
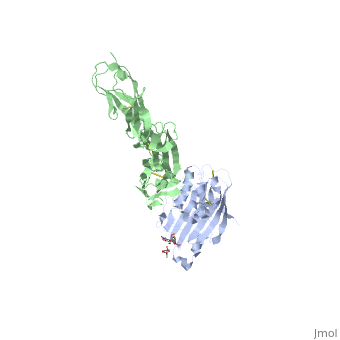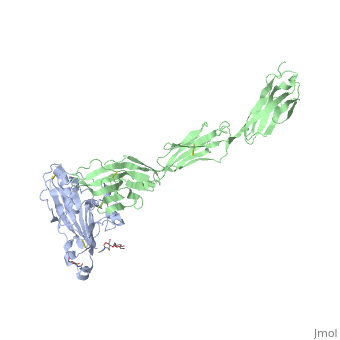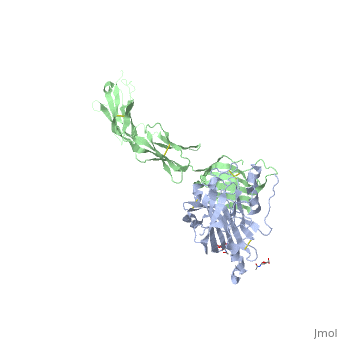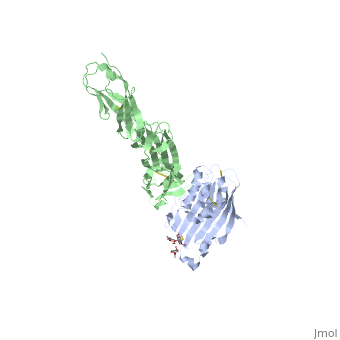
| | ==Structure of HSV-2 gD bound to nectin-1== | | ==Structure of HSV-2 gD bound to nectin-1== |
| - | <StructureSection load='4myw' size='340' side='right' caption='[[4myw]], [[Resolution|resolution]] 3.19Å' scene=''> | + | <StructureSection load='4myw' size='340' side='right'caption='[[4myw]], [[Resolution|resolution]] 3.19Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[4myw]] is a 4 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4MYW OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4MYW FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[4myw]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Human_herpesvirus_2_strain_333 Human herpesvirus 2 strain 333]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4MYW OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4MYW FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.189Å</td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3u82|3u82]], [[4myv|4myv]]</td></tr> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4myw FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4myw OCA], [http://pdbe.org/4myw PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=4myw RCSB], [http://www.ebi.ac.uk/pdbsum/4myw PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=4myw ProSAT]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4myw FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4myw OCA], [https://pdbe.org/4myw PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4myw RCSB], [https://www.ebi.ac.uk/pdbsum/4myw PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4myw ProSAT]</span></td></tr> |
| | </table> | | </table> |
| - | == Disease == | |
| - | [[http://www.uniprot.org/uniprot/PVRL1_HUMAN PVRL1_HUMAN]] Zlotogora-Ogur syndrome;Cleft lip with or without cleft palate. The disease is caused by mutations affecting the gene represented in this entry. The disease is caused by mutations affecting the gene represented in this entry. | |
| | == Function == | | == Function == |
| - | [[http://www.uniprot.org/uniprot/GD_HHV23 GD_HHV23]] Envelope glycoprotein that binds to the potential host cell entry receptors TNFRSF14/HVEM, PVRL1 and PVRL1. May trigger fusion with host membrane, by recruiting the fusion machinery composed of gB and gH/gL (By similarity). [[http://www.uniprot.org/uniprot/PVRL1_HUMAN PVRL1_HUMAN]] Promotes cell-cell contacts by forming homophilic or heterophilic trans-dimers. Heterophilic interactions have been detected between PVRL1/nectin-1 and PVRL3/nectin-3 and between PVRL1/nectin-1 and PVRL4/nectin-4. Functions as an entry receptor for herpes simplex virus and pseudorabies virus.<ref>PMID:7721102</ref> | + | [https://www.uniprot.org/uniprot/GD_HHV23 GD_HHV23] Envelope glycoprotein that binds to the potential host cell entry receptors TNFRSF14/HVEM, PVRL1 and PVRL1. May trigger fusion with host membrane, by recruiting the fusion machinery composed of gB and gH/gL (By similarity). |
| | <div style="background-color:#fffaf0;"> | | <div style="background-color:#fffaf0;"> |
| | == Publication Abstract from PubMed == | | == Publication Abstract from PubMed == |
| Line 24: |
Line 22: |
| | ==See Also== | | ==See Also== |
| | *[[Glycoproteins B and D|Glycoproteins B and D]] | | *[[Glycoproteins B and D|Glycoproteins B and D]] |
| | + | *[[Poliovirus receptor-related protein|Poliovirus receptor-related protein]] |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Chen, Z]] | + | [[Category: Homo sapiens]] |
| - | [[Category: Gao, G F]] | + | [[Category: Human herpesvirus 2 strain 333]] |
| - | [[Category: Li, Y]] | + | [[Category: Large Structures]] |
| - | [[Category: Lu, G]] | + | [[Category: Chen Z]] |
| - | [[Category: Qi, J]] | + | [[Category: Gao GF]] |
| - | [[Category: Yan, J]] | + | [[Category: Li Y]] |
| - | [[Category: Zhang, N]] | + | [[Category: Lu G]] |
| - | [[Category: Zheng, C]] | + | [[Category: Qi J]] |
| - | [[Category: Cell surface attachment]] | + | [[Category: Yan J]] |
| - | [[Category: Cellular surface]]
| + | [[Category: Zhang N]] |
| - | [[Category: Igv-like core]]
| + | [[Category: Zheng C]] |
| - | [[Category: N-/c-terminal extension]] | + | |
| - | [[Category: Receptor binding]] | + | |
| - | [[Category: Tandem arranged v-c-c ig-like domain]]
| + | |
| - | [[Category: Viral]]
| + | |
| - | [[Category: Viral protein-cell adhesion complex]]
| + | |
| Structural highlights
Function
GD_HHV23 Envelope glycoprotein that binds to the potential host cell entry receptors TNFRSF14/HVEM, PVRL1 and PVRL1. May trigger fusion with host membrane, by recruiting the fusion machinery composed of gB and gH/gL (By similarity).
Publication Abstract from PubMed
Herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) are among the most prevalent human pathogens. Both viruses can recognize, via the surface envelope glycoprotein D (gD), human nectin-1 as a functional receptor. Previous studies have successfully elucidated the molecular basis of the binding between HSV-1 gD and nectin-1 by co-crystallography. Despite a high sequence identity between HSV-1 and -2 gDs, the atomic inter-molecule details for HSV-2-gD/nectin-1 interaction remain elusive. Here, we reported the crystal structures of both the unbound and the nectin-1-bound HSV-2 gD. The free gD structure expectedly comprises an IgV-like core and the surface-exposed terminal extensions as observed in its HSV-1 counterpart, but lacks traceable electron densities for a large portion of the terminal elements. These terminal residues were clearly traced in the complex structure as a definitive loop in the N-terminus and an alpha-helix in the C-terminus, thereby showing a conserved nectin-1-binding mode as reported for HSV-1 gD. The interface residues in nectin-1 were further mutated and tested for the gD-interaction by surface plasmon resonance. The resultant binding patterns were similar between HSV-1 and -2 gDs, further supporting a homologous receptor-binding basis by the two viruses for nectin-1. These data, together with a cell-based fusion assay showing a cross-inhibition of the gD/nectin-1 mediated cell-cell fusion by soluble HSV-1 and -2 gDs, provided solid structural and functional evidence that HSV-1/-2 recognizes nectin-1 via the same binding mode. Finally, we also demonstrated that nectin-1 I80 is an important residue involved in gD interaction. IMPORTANCE: Despite intensified studies, a detailed picture of the molecular features in the HSV-2-gD/nectin-1 interaction remains unavailable. Previous work focused on HSV-1 gD, which folds into an IgV-like core with large terminal extensions and utilizes the extension elements to engage nectin-1. Here, we reported the crystal structures of HSV-2 gD in both the free and the nectin-1-bound forms. The atomic inter-molecule details for HSV-2-gD/nectin-1 interaction were clearly presented. The observed binding mode is identical to that reported for its HSV-1 counterpart. This structural observation was further supported by our comparative functional assays showing that nectin-1 mutations similarly affect the ligand/receptor interaction of both virus gDs. Taken together, we provided comprehensive structural and functional data demonstrating a conserved receptor-binding mode between HSV-1 and -2 for nectin-1. Our results also indicate that the tropism-difference between the two viruses likely arises from other aspects rather than the gD/nectin-1 binding-features.
Crystal structure of HSV-2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.,Lu G, Zhang N, Qi J, Li Y, Chen Z, Zheng C, Gao GF, Yan J J Virol. 2014 Sep 17. pii: JVI.01906-14. PMID:25231300[1]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Lu G, Zhang N, Qi J, Li Y, Chen Z, Zheng C, Gao GF, Yan J. Crystal structure of HSV-2 gD bound to nectin-1 reveals a conserved mode of receptor recognition. J Virol. 2014 Sep 17. pii: JVI.01906-14. PMID:25231300 doi:http://dx.doi.org/10.1128/JVI.01906-14
|