Aminomutase
From Proteopedia
(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load=' | + | <StructureSection load='' size='340' side='right' caption='Tyrosine aminomutase complex with peptide-derived chromophore cofactor (PDB code [[2ohy]])' scene='71/715468/Cv/1'> |
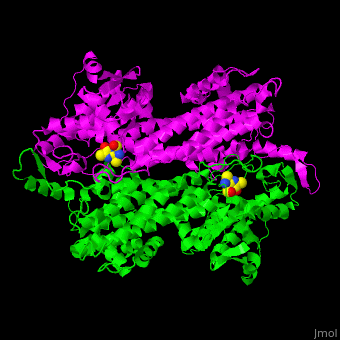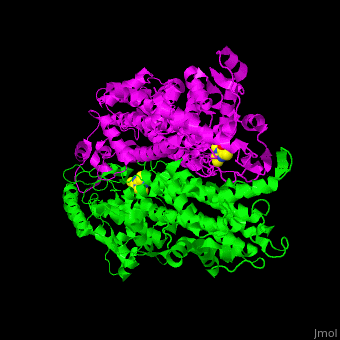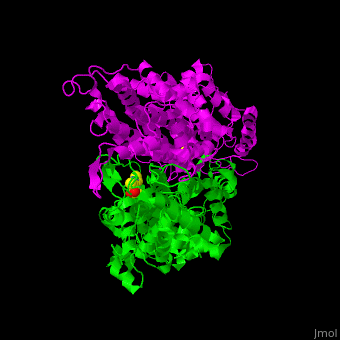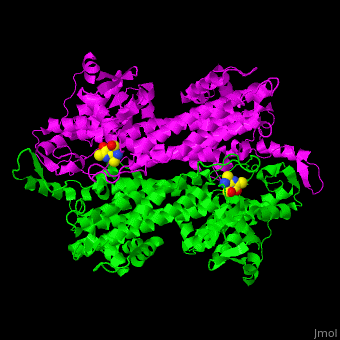
'''Aminomutases''' catalyze the exchange of hydrogen atom and an amine substituent on a neighboring carbon atom.<ref name="Ad">PMID:17516659</ref> | '''Aminomutases''' catalyze the exchange of hydrogen atom and an amine substituent on a neighboring carbon atom.<ref name="Ad">PMID:17516659</ref> | ||
| Line 32: | Line 32: | ||
== Structural highlights == | == Structural highlights == | ||
| - | TAM active site residues facilitate the deprotonation/reprotonation reaction. <scene name='71/715468/Cv/ | + | TAM active site residues facilitate the deprotonation/reprotonation reaction. <scene name='71/715468/Cv/3'>The active site</scene> includes residues from both monomers in Tyrosine aminomutase (PDB code [[2ohy]]).<ref name="Ad">PMID:17516659</ref> |
| - | + | ||
| - | + | ||
==3D structures of aminomutase== | ==3D structures of aminomutase== | ||
| + | [[Aminomutase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[2zsl]], [[2zsm]], [[2epj]] – GSAM + PLP derivative – ''Aeropyrum pernix''<br /> | ||
| - | **[[2hp1]], [[4gsa]] – SeGSAM + PLP <br /> | ||
| - | **[[2hp2]] – SeGSAM + PLP + aminopentanoic acid derivative<br /> | ||
| - | **[[2cfb]], [[2hoz]], [[3fq7]] – SeGSAM + PLP derivative <br /> | ||
| - | **[[3fq8]] – SeGSAM (mutant) + PLP derivative <br /> | ||
| - | **[[2gsa]] – SeGSAM + PLP derivative + PLP<br /> | ||
| - | **[[2gsb]], [[3fqa]] – SeGSAM + PLP derivative + gabaculine<br /> | ||
| - | **[[2e7u]] – GSAM + PLP derivative – ''Thermus thermophilus''<br /> | ||
| - | **[[3bs8]] – GSAM (mutant) + PLP derivative – ''Bacillus subtilis''<br /> | ||
| - | **[[3l44]] – BaGSAM + PLP derivative – ''Bacillus anthracis''<br /> | ||
| - | **[[3k28]] – BaGSAM + PLP <br /> | ||
| - | **[[5hdm]] – GSAM + PLP + PMP - ''Arabidopsis thaliana'' <br /> | ||
| - | **[[3k28]] – BaGSAM + PLP <br /> | ||
| - | **[[5i92]] – GSAM + leucine - ''Pseudomonas aeruginosa'' <br /> | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Christianson CV, Montavon TJ, Van Lanen SG, Shen B, Bruner SD. The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry. 2007 Jun 19;46(24):7205-14. Epub 2007 May 22. PMID:17516659 doi:10.1021/bi7003685