|
|
| (41 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | <StructureSection load='3hjh' size='350' side='right' scene='' caption='Transcription-repair coupling factor complex with Co+2 ion (red), [[3hjh]]'>
| + | __TOC__ |
| - | [[Image:2eyq.png|left|200px|thumb|Crystal Structure of Transcription-repair coupling factor [[2eyq]]]]
| + | The bacterial [[transcription-repair coupling factor]] TRCF, also called Mfd translocase, is a DNA repair protein. It has a role in transcription-coupled repair, a subpathway of nucleotide excision repair (NER). Mfd recognizes stalled RNA polymerase (RNAP) and either restarts transcription or removes the stalled polymerase and recruits the NER proteins UvrA and UvrB. |
| | | | |
| | + | == Function == |
| | + | Mfd has ATP hydrolysis activity, DNA binding sites and a UvrA binding sites. These three functions are inhibited in the isolated enzyme, but are activated when Mfd encounters stalled RNA polymerase (or through various mutations that remove inhibitory domains <ref>PMID: 27864884</ref>). Mfd also contains an RNA interaction domain (RID) that binds to the beta subunit of RNAP. |
| | | | |
| | + | == Relationship to other enzymes == |
| | + | The N-terminal part of Mfd shows sequence similarity to UvrB, including in the domain of UvrB that interacts with UvrA. However, the conserved helicase motifs present in UvrB (responsible for binding and hydrolyzing ATP) are absent in that part of Mfd. Furthermore, the sequence segment known to fold as a beta hairpin in UvrB (involved in clamping down a single strand of DNA) seems absent. The C-terminal part of Mfd shows sequence similarity to SF1/SF2 helicases (UvrB is an example), containing conserved helicase/translocase motifs. |
| | | | |
| | + | == Structure and conformational change == |
| | + | <StructureSection load='' size='350' side='right' scene='2eyq/Domainscolorsflabe/3' caption=''> |
| | + | The initial scene (switch to <scene name='46/460252/Apo/1'>cartoon</scene>) shows the domains of Mfd in the conformation of the apo-enzyme.<ref>DOI:10.1016/j.cell.2005.11.045</ref> The UvrA interaction site (on domain 2) is occluded by domain 7. The translocase domains (domain 5 and domain 6), through interactions with domains 1 and 3, are locked in an inactive conformation, preventing the typical hinge motion of translocases when they bind and hydrolyze ATP while moving along DNA. When Mfd binds to DNA in the presence of ADP·AlFₓ, Mfd undergoes large domain rearrangements, giving a hint how it might bind to stalled RNAP.<ref>DOI:10.1038/s41467-020-17457-1</ref> A first glimpse of the Mfd RNAP complex was through cryo-EM, but large parts of the Mfd protein were not resolved.<ref>DOI:10.1093/nar/gkaa904</ref> |
| | | | |
| | + | [[Image:Mfd domains.JPG|thumb|left]] |
| | | | |
| | + | In 2021, the structures of several intermediates in the presences of stalled RNAP were determined using cryo-EM, shedding further light on the loading and activation mechanism, as well as how translocation of Mfd on DNA leads to disruption of the RNAP elongation complex and recruitment of UvrA.<ref>doi: 10.7554/eLife.62117</ref> |
| | + | When <scene name='46/460252/Mfd_rnap_l1/2'>Mfd binds to a stalled RNA polymerase</scene>, the RID (domain 4) binds to the beta subunit of RNA polymerase and domains 5 and domain 6 bind to DNA and ATP. These multiple binding events require a different relative orientation of RID and the translocase domains, and the necessary conformational changes disrupt inter-domain interactions seen in the apo-structure while activating the translocase activity of Mfd. |
| | | | |
| | + | Once the translocase domains are activated, they move closer and closer to the surface of the RNA polymerase in multiple cycles of ATP hydrolysis until <scene name='46/460252/Mfd_rnap_l2/2'>they make contact</scene>. The translocase moving along the DNA together with the RID tethered at a fixed point on the RNAP surface result in substantial reorganization of the domain contacts and relative orientation. Eventually, this leads to the the <scene name='46/460252/Mfd_rnap_c4/1'>UvrA interaction site on domain 2 (cyan) becoming available for UvrA</scene>, thus recruiting the NER machinery to the damaged template strand for subsequent repair. Further translocation (not shown) will lead to distabilization of the RNAP: nucleic acid complex, helping to dislodge the stalled RNAP. The changing domain contacts are schematically summarized below. |
| | | | |
| | + | [[Image:Mfd domain contacts.JPG|400px|thumb|center|Distinct domain contacts in various Mfd structures]] |
| | | | |
| | + | One of the functions of the large conformational change is to create a topological lock around the DNA to support translocation in a highly processive manner. Whereas in other systems, motor protein, clamp and clamp loader are separate proteins, all these functions are housed here in a single polypeptide. Mfd uses a single ATPase activity to create a sliding clamp and load it onto the DNA. The necessary reorganization of domains is visualized in a <scene name='46/460252/Mfd_domain_contacts/2'>morph</scene>. <ref>The [[Jmol/Storymorph|Storymorph Jmol scripts]] were used to create the interpolation shown in the morph. [https://proteopedia.org/wiki/index.php/Image:Morph_mfd_contacts.pdb Coordinates] available on Proteopedia</ref> |
| | + | {{Template:Button Toggle Animation2}} |
| | | | |
| - | | + | In the apo-structure, domains 3 and 4 are far apart from each other (connected by an extended linker), but they make direct contacts when Mfd is loaded on DNA. On the other hand, domains 4 and 5 make direct contacts in the apo-structure, but are far apart from each other (coonected by an extended linker) when Mfd is loaded on DNA. Finally, domain 2 and domain 7 bind to each other in the apo-state whereas domain 2 is available for binding UvrA in the DNA-bound state. |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | == Function ==
| + | |
| - | '''Transcription-repair coupling factor (TRCF)''' enables the coupling of these processes in bacteria and humans. The TRCF has ATPase activity. <scene name='2eyq/Asymmunit/2'>The crystallographic asymmetric unit of the solved structure contains 2 monomers of Mfd (residues 2-1147 of 1151 for one molecule and 5-1147 of the other), five HEPES molecules, and the sulfate ions.</scene>
| + | |
| - | | + | |
| - | Size exclusion chromatogaphy indicates that the protein <scene name='2eyq/Biolunit/1'>exists as a monomer in solution.</scene>
| + | |
| - | == Structural highlights ==
| + | |
| - | <scene name='2eyq/Figonea/2'>The monomer colored as</scene> an N→C rainbow similar to [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig1&_ba=1&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=a4326e4023d32f5de977da04b9a9fdf6 Figure 1A] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure. {{Link Toggle FancyCartoonHighQualityView}}.
| + | |
| - | | + | |
| - | The domains in the structure of Mfd:
| + | |
| - | *<scene name='2eyq/Domain1a/2'>D1a</scene>
| + | |
| - | *<scene name='2eyq/Domaind2/2'>D2 (UvrB homology module)</scene>
| + | |
| - | *<scene name='2eyq/Domain1b/2'>D1b</scene>
| + | |
| - | *<scene name='2eyq/Domaind3/2'>D3</scene>
| + | |
| - | *<scene name='2eyq/Domain4/2'>D4 (RNAP Interacting Domain)</scene>
| + | |
| - | *<scene name='2eyq/Translocation_module/3'>Translocation module (D5 and D6)</scene>
| + | |
| - | *<scene name='2eyq/Domain7/2'>D7 (handle)</scene>
| + | |
| - | <scene name='2eyq/Domainscoloredlabeled/1'>The domains all shown and colored</scene> similar to [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig1&_ba=1&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=a4326e4023d32f5de977da04b9a9fdf6 Figure 1B] and [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig2&_ba=2&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b56879d585430e1112f6c5c081a1d158 Figure 2] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure.
| + | |
| - | | + | |
| - | <scene name='2eyq/Domainscolorsflabe/3'>Domains labeled and colored and displayed similar to</scene> [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig1&_ba=1&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=a4326e4023d32f5de977da04b9a9fdf6 Figure 1B] and [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig2&_ba=2&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b56879d585430e1112f6c5c081a1d158 Figure 2] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure. <scene name='2eyq/Domainscolorspacefill/1'>Unlabeled</scene>.<br>
| + | |
| - | <nowiki>[</nowiki>Note: the following view generates a substantial surface which may take several minutes to calculate. Use the one above as an alternative unless you are willing to spend the time.<nowiki>]</nowiki> <scene name='2eyq/Domainssurflabel/1'>Domains labeled and represented much more similar to</scene> [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig1&_ba=1&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=a4326e4023d32f5de977da04b9a9fdf6 Figure 1B]. {{Link Toggle LabelsOff}}.
| + | |
| - | | + | |
| - | <scene name='2eyq/Translocationmodcontext/1'>The translocation module shown in the context of the Mfd</scene> displayed as in
| + | |
| - | [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig3&_ba=3&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=580f83ffa975d90b5c9d9dc0eb930d8c Figure 3A] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure.
| + | |
| - | | + | |
| - | <scene name='2eyq/Translocation_modhelciasemotif/1'>The seven ATPase/helicase motifs in the translocation domain</scene> are highlighted in red. See figure 3B.
| + | |
| - | | + | |
| - | <scene name='2eyq/Walkera/3'>Walker A motif of Mfd</scene> displayed similar to
| + | |
| - | [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig3&_ba=3&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=580f83ffa975d90b5c9d9dc0eb930d8c Figure 3B] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure.
| + | |
| - | | + | |
| - | <scene name='2eyq/Figure3c/3'>TRG motif and motif VI</scene> indicated similar to [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig3&_ba=3&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=580f83ffa975d90b5c9d9dc0eb930d8c Figure 3C] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure.
| + | |
| | | | |
| | == 3D Structures of Transcription-repair coupling factor == | | == 3D Structures of Transcription-repair coupling factor == |
| | + | [[Transcription-repair coupling factor 3D structures]] |
| | | | |
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}}
| + | == References == |
| | + | <references/> |
| | | | |
| - | [[2eyq]], [[2b2n]] – EcTRCF – ''Escherichia coli''<br /> | |
| - | [[3hjh]] – EcTRCF residues 1-470<br /> | |
| - | [[4dfc]] – EcTRCF D2 domain + UvrABC system protein A<br /> | |
| - | [[3mlq]] – TRCF RNA polymerase interacting domain + DNA-directed RNA polymerase subunit β - ''Thermus thermophilus''<br /> | |
| - | [[2qsr]] – TRCF C terminal – ''Streptococcus pneumoni''<br /> | |
| - | </StructureSection> | |
| - | | |
| - | ==Reference== | |
| - | <ref group="xtra">PMID:16469698</ref><references group="xtra"/> | |
| | [[Category: Escherichia coli]] | | [[Category: Escherichia coli]] |
| | [[Category: Darst, S A.]] | | [[Category: Darst, S A.]] |
| Line 85: |
Line 48: |
| | [[Category:Topic Page]] | | [[Category:Topic Page]] |
| | <br /> | | <br /> |
| | + | </StructureSection> |
| | Created with the participation of [[User:Wayne Decatur|Wayne Decatur]] | | Created with the participation of [[User:Wayne Decatur|Wayne Decatur]] |
Mfd has ATP hydrolysis activity, DNA binding sites and a UvrA binding sites. These three functions are inhibited in the isolated enzyme, but are activated when Mfd encounters stalled RNA polymerase (or through various mutations that remove inhibitory domains [1]). Mfd also contains an RNA interaction domain (RID) that binds to the beta subunit of RNAP.
The N-terminal part of Mfd shows sequence similarity to UvrB, including in the domain of UvrB that interacts with UvrA. However, the conserved helicase motifs present in UvrB (responsible for binding and hydrolyzing ATP) are absent in that part of Mfd. Furthermore, the sequence segment known to fold as a beta hairpin in UvrB (involved in clamping down a single strand of DNA) seems absent. The C-terminal part of Mfd shows sequence similarity to SF1/SF2 helicases (UvrB is an example), containing conserved helicase/translocase motifs.
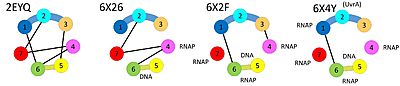
| The initial scene (switch to ) shows the domains of Mfd in the conformation of the apo-enzyme.[2] The UvrA interaction site (on domain 2) is occluded by domain 7. The translocase domains (domain 5 and domain 6), through interactions with domains 1 and 3, are locked in an inactive conformation, preventing the typical hinge motion of translocases when they bind and hydrolyze ATP while moving along DNA. When Mfd binds to DNA in the presence of ADP·AlFₓ, Mfd undergoes large domain rearrangements, giving a hint how it might bind to stalled RNAP.[3] A first glimpse of the Mfd RNAP complex was through cryo-EM, but large parts of the Mfd protein were not resolved.[4]
In 2021, the structures of several intermediates in the presences of stalled RNAP were determined using cryo-EM, shedding further light on the loading and activation mechanism, as well as how translocation of Mfd on DNA leads to disruption of the RNAP elongation complex and recruitment of UvrA.[5]
When , the RID (domain 4) binds to the beta subunit of RNA polymerase and domains 5 and domain 6 bind to DNA and ATP. These multiple binding events require a different relative orientation of RID and the translocase domains, and the necessary conformational changes disrupt inter-domain interactions seen in the apo-structure while activating the translocase activity of Mfd.
Once the translocase domains are activated, they move closer and closer to the surface of the RNA polymerase in multiple cycles of ATP hydrolysis until . The translocase moving along the DNA together with the RID tethered at a fixed point on the RNAP surface result in substantial reorganization of the domain contacts and relative orientation. Eventually, this leads to the the , thus recruiting the NER machinery to the damaged template strand for subsequent repair. Further translocation (not shown) will lead to distabilization of the RNAP: nucleic acid complex, helping to dislodge the stalled RNAP. The changing domain contacts are schematically summarized below.
 Distinct domain contacts in various Mfd structures One of the functions of the large conformational change is to create a topological lock around the DNA to support translocation in a highly processive manner. Whereas in other systems, motor protein, clamp and clamp loader are separate proteins, all these functions are housed here in a single polypeptide. Mfd uses a single ATPase activity to create a sliding clamp and load it onto the DNA. The necessary reorganization of domains is visualized in a . [6]
In the apo-structure, domains 3 and 4 are far apart from each other (connected by an extended linker), but they make direct contacts when Mfd is loaded on DNA. On the other hand, domains 4 and 5 make direct contacts in the apo-structure, but are far apart from each other (coonected by an extended linker) when Mfd is loaded on DNA. Finally, domain 2 and domain 7 bind to each other in the apo-state whereas domain 2 is available for binding UvrA in the DNA-bound state.
3D Structures of Transcription-repair coupling factor
Transcription-repair coupling factor 3D structures
References
- ↑ Selby CP. Mfd Protein and Transcription-Repair Coupling in Escherichia coli. Photochem Photobiol. 2017 Jan;93(1):280-295. doi: 10.1111/php.12675. Epub 2017, Jan 18. PMID:27864884 doi:http://dx.doi.org/10.1111/php.12675
- ↑ Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006 Feb 10;124(3):507-20. PMID:16469698 doi:10.1016/j.cell.2005.11.045
- ↑ Brugger C, Zhang C, Suhanovsky MM, Kim DD, Sinclair AN, Lyumkis D, Deaconescu AM. Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd. Nat Commun. 2020 Jul 27;11(1):3740. doi: 10.1038/s41467-020-17457-1. PMID:32719356 doi:http://dx.doi.org/10.1038/s41467-020-17457-1
- ↑ Shi J, Wen A, Zhao M, Jin S, You L, Shi Y, Dong S, Hua X, Zhang Y, Feng Y. Structural basis of Mfd-dependent transcription termination. Nucleic Acids Res. 2020 Nov 18;48(20):11762-11772. doi: 10.1093/nar/gkaa904. PMID:33068413 doi:http://dx.doi.org/10.1093/nar/gkaa904
- ↑ Kang JY, Llewellyn E, Chen J, Olinares PDB, Brewer J, Chait BT, Campbell EA, Darst SA. Structural basis for transcription complex disruption by the Mfd translocase. Elife. 2021 Jan 22;10. pii: 62117. doi: 10.7554/eLife.62117. PMID:33480355 doi:http://dx.doi.org/10.7554/eLife.62117
- ↑ The Storymorph Jmol scripts were used to create the interpolation shown in the morph. Coordinates available on Proteopedia
|


