We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Megan Harris./Sandbox 1
From Proteopedia
< User:Megan Harris.(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='5EPY' size='340' side='right' caption=' | + | [http://www.example.com link title]<scene name='74/745998/Grazoprevir/3'>Text To Be Displayed</scene><StructureSection load='5EPY' size='340' side='right' caption='Zepatier: A combination drug composed of Elbasvir and Grazoprevir' scene=''> |
| Line 10: | Line 10: | ||
== Structure and Mechanism of Elbasvir == | == Structure and Mechanism of Elbasvir == | ||
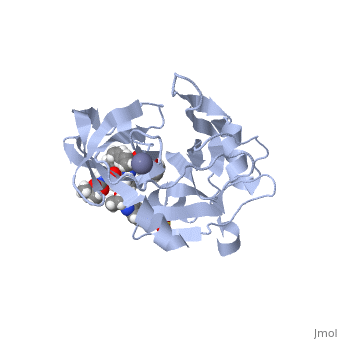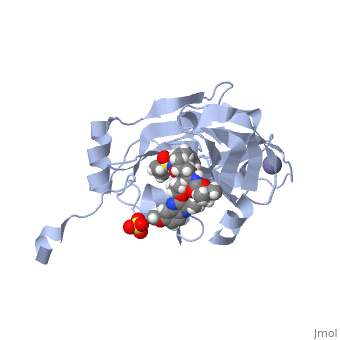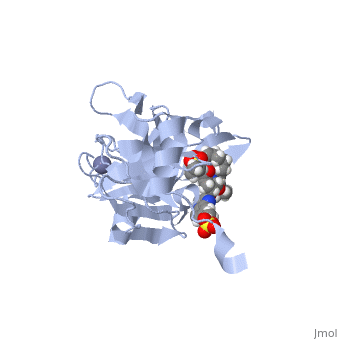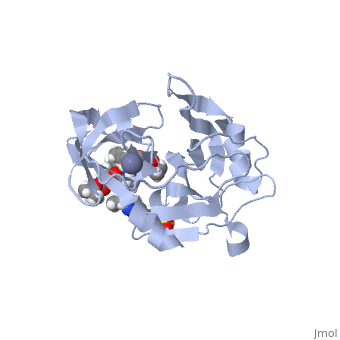
| - | Elbasvir, MK-8742,a tetra-cyclic indole based NS5A inhibitor, was experimentally obtained through the modification of MK-4882. MK-4882 was one of the first clinical candidate drugs as an NS5A inhibitor. Although initially effective, viral breakthrough was a concern which led to continue development of compounds that would be effective against multiple genotypes as well as NS5A mutations. Through efficacy studies of infected chimpanzees, Elbasvir was discovered as a diastereomer of the known compound, MK-4882 <ref>DOI: 10.1002/cmdc.201300343</ref>. Elbasvir has a structural formula of C<sub>49</sub>H<sub>55</sub>N<sub>9</sub>O<sub>7</sub> and a molecular weight of 882.035 g/mol. It is a highly flexible protein with 13 rotatable bonds. It has four hydrogen bond donors and nine hydrogen bond acceptors <ref>National Center for Biotechnology Inforamtion. PubChem Compound Database; CID=71661251, https://pubchem.ncbi.nlm.nih.gov/compound/71661251#section=Chemical-and-Physical-Properties</ref>. | + | <scene name='74/745998/Elbasvir_structure1/7'>Elbasvir</scene>, MK-8742,a tetra-cyclic indole based NS5A inhibitor, was experimentally obtained through the modification of MK-4882. MK-4882 was one of the first clinical candidate drugs as an NS5A inhibitor. Although initially effective, viral breakthrough was a concern which led to continue development of compounds that would be effective against multiple genotypes as well as NS5A mutations. Through efficacy studies of infected chimpanzees, Elbasvir was discovered as a diastereomer of the known compound, MK-4882 <ref>DOI: 10.1002/cmdc.201300343</ref>. Elbasvir has a structural formula of C<sub>49</sub>H<sub>55</sub>N<sub>9</sub>O<sub>7</sub> and a molecular weight of 882.035 g/mol. It is a highly flexible protein with 13 rotatable bonds. It has four hydrogen bond donors and nine hydrogen bond acceptors <ref>National Center for Biotechnology Inforamtion. PubChem Compound Database; CID=71661251, https://pubchem.ncbi.nlm.nih.gov/compound/71661251#section=Chemical-and-Physical-Properties</ref>. |
Although the actual mechanism of Elbasvir as a NS5A inhibitor is unknown, several hypothetical mechanisms have been proposed. The NS5A protein is a critical protein for both DNA replication and assembly in the hepatitis C virus. With critical roles in both processes, NS5A is an excellent source as a target for inhibitors. NS5A is naturally located in the endoplasmic reticulum of the cell. When inhibited, the protein is redistributed from the ER to lipid droplets. If the cellular location changed, NS5A would be unable to aid in viral replication, illustrating a form of protein inhibition that Elbasvir could cause. Another potential mechanism of NS5A inhibitors is the altering of the phosphorylation of the NS5A protein <ref>DOI: http://dx.doi.org/10.1016/j.jhep.2013.03.030</ref>. The function of the NS5A protein is highly dependent on the phosphorylation, both basal phosphorylation and hyperphosphorylation <ref>doi: 10.1128/JVI.00253-11</ref>. Since the phosphorylation of the NS5A protein is critical for protein function, altering the levels of phosphorylation could impact the activity of the protein. | Although the actual mechanism of Elbasvir as a NS5A inhibitor is unknown, several hypothetical mechanisms have been proposed. The NS5A protein is a critical protein for both DNA replication and assembly in the hepatitis C virus. With critical roles in both processes, NS5A is an excellent source as a target for inhibitors. NS5A is naturally located in the endoplasmic reticulum of the cell. When inhibited, the protein is redistributed from the ER to lipid droplets. If the cellular location changed, NS5A would be unable to aid in viral replication, illustrating a form of protein inhibition that Elbasvir could cause. Another potential mechanism of NS5A inhibitors is the altering of the phosphorylation of the NS5A protein <ref>DOI: http://dx.doi.org/10.1016/j.jhep.2013.03.030</ref>. The function of the NS5A protein is highly dependent on the phosphorylation, both basal phosphorylation and hyperphosphorylation <ref>doi: 10.1128/JVI.00253-11</ref>. Since the phosphorylation of the NS5A protein is critical for protein function, altering the levels of phosphorylation could impact the activity of the protein. | ||
| - | == Mechanism of Grazoprevir == | + | == Structure and Mechanism of Grazoprevir == |
| - | Grazoprevir stops the hepatitis C virus by inhibiting the NS3/4A protease, which is an enzyme responsible for the cleavage of the viral polyprotein. The molecular formula is C<sub>38</sub>H<sub>50</sub>N<sub>6</sub>O<sub>9</sub>S and a molecular weight of 766.911 g/mol <ref>National Center for Biotechnology Information. PubChem Compound Database; CID=44603531, https://pubchem.ncbi.nlm.nih.gov/compound/44603531.</ref>. Cleavage of this polyprotein facilitates viral assembly and maturation of the virus. The NS3/4A protease is a protein complex in which the active site consists of a catalytic triad of amino acids S139, H57 and D81, and an activating cofactor which assists in, and is required for the activity of the protease. Grazoprevir has been proven to be more effective than similar protease inhibitors due to its two-way approach to inhibition. NS3/4A protease cleaves both the viral polyprotein of the Hep C virus and host factors involved in the immune response, such as TRIF and MAVS. Grazoprevir is unique in regard to its P4 cap, and P2 to P4 macrocyclic restriction, which interacts with the catalytic triad of the NS3/4A protease in its own distinct conformation. In this conformation, the P2 region is comprised of the Histidine-57 and Aspartate-81 residues. | + | <scene name='74/745998/Grazoprevir/3'>Grazoprevir</scene> stops the hepatitis C virus by inhibiting the NS3/4A protease, which is an enzyme responsible for the cleavage of the viral polyprotein. The molecular formula is C<sub>38</sub>H<sub>50</sub>N<sub>6</sub>O<sub>9</sub>S and a molecular weight of 766.911 g/mol <ref>National Center for Biotechnology Information. PubChem Compound Database; CID=44603531, https://pubchem.ncbi.nlm.nih.gov/compound/44603531.</ref>. Cleavage of this polyprotein facilitates viral assembly and maturation of the virus. The NS3/4A protease is a protein complex in which the active site consists of a catalytic triad of amino acids S139, H57 and D81, and an activating cofactor which assists in, and is required for the activity of the protease. Grazoprevir has been proven to be more effective than similar protease inhibitors due to its two-way approach to inhibition. NS3/4A protease cleaves both the viral polyprotein of the Hep C virus and host factors involved in the immune response, such as TRIF and MAVS. Grazoprevir is unique in regard to its P4 cap, and P2 to P4 macrocyclic restriction, which interacts with the catalytic triad of the NS3/4A protease in its own distinct conformation. In this conformation, the P2 region is comprised of the Histidine-57 and Aspartate-81 residues. |
| - | == Disease == | + | == Disease in Humans == |
Hepatitis C, hereafter known as HCV, is a viral infection that causes inflammation of the liver and decreasing liver functioning. First isolated by Michael Houghton in 1989, hepatitis C was discovered to encode a single polyprotein of 3,000 amino acids later cleaved into 10 polypeptides. Polypeptides produced include p7 ion channel proteins, the core protein, glycoproteins envelope 1 (E1) and envelope 2 (E2), and nonstructural proteins NS1, NS2, NS3, NS4A, NS5A, and NS5B. The 5’ untranslated region of HCV is not capped, allowing it to fold into a complex secondary RNA structure hereby creating an internal ribosome entry site that is able to direct ribosomal subunits and cellular factors, and as a result, translation. Development of cell culture based model systems have allowed for the study of the HCV lifecycle <ref>Chevaliez, S.; Pawlotsky, J. M. Virology of hepatitis C virus infection. Best Pract. Res. Clin. Gastroenterol.2012, 26, 381-389.</ref>. | Hepatitis C, hereafter known as HCV, is a viral infection that causes inflammation of the liver and decreasing liver functioning. First isolated by Michael Houghton in 1989, hepatitis C was discovered to encode a single polyprotein of 3,000 amino acids later cleaved into 10 polypeptides. Polypeptides produced include p7 ion channel proteins, the core protein, glycoproteins envelope 1 (E1) and envelope 2 (E2), and nonstructural proteins NS1, NS2, NS3, NS4A, NS5A, and NS5B. The 5’ untranslated region of HCV is not capped, allowing it to fold into a complex secondary RNA structure hereby creating an internal ribosome entry site that is able to direct ribosomal subunits and cellular factors, and as a result, translation. Development of cell culture based model systems have allowed for the study of the HCV lifecycle <ref>Chevaliez, S.; Pawlotsky, J. M. Virology of hepatitis C virus infection. Best Pract. Res. Clin. Gastroenterol.2012, 26, 381-389.</ref>. | ||
Current revision
link title
| |||||||||||
References
- ↑ Soumana DI, Kurt Yilmaz N, Prachanronarong KL, Aydin C, Ali A, Schiffer CA. Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172. ACS Chem Biol. 2015 Dec 18. PMID:26682473 doi:http://dx.doi.org/10.1021/acschembio.5b00647
- ↑ Coburn CA, Meinke PT, Chang W, Fandozzi CM, Graham DJ, Hu B, Huang Q, Kargman S, Kozlowski J, Liu R, McCauley JA, Nomeir AA, Soll RM, Vacca JP, Wang D, Wu H, Zhong B, Olsen DB, Ludmerer SW. Discovery of MK-8742: an HCV NS5A inhibitor with broad genotype activity. ChemMedChem. 2013 Dec;8(12):1930-40. doi: 10.1002/cmdc.201300343. Epub 2013 Oct, 14. PMID:24127258 doi:http://dx.doi.org/10.1002/cmdc.201300343
- ↑ National Center for Biotechnology Inforamtion. PubChem Compound Database; CID=71661251, https://pubchem.ncbi.nlm.nih.gov/compound/71661251#section=Chemical-and-Physical-Properties
- ↑ doi: https://dx.doi.org/http
- ↑ Fridell RA, Qiu D, Valera L, Wang C, Rose RE, Gao M. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J Virol. 2011 Jul;85(14):7312-20. doi: 10.1128/JVI.00253-11. Epub 2011 May 18. PMID:21593143 doi:http://dx.doi.org/10.1128/JVI.00253-11
- ↑ National Center for Biotechnology Information. PubChem Compound Database; CID=44603531, https://pubchem.ncbi.nlm.nih.gov/compound/44603531.
- ↑ Chevaliez, S.; Pawlotsky, J. M. Virology of hepatitis C virus infection. Best Pract. Res. Clin. Gastroenterol.2012, 26, 381-389.
- ↑ Petrescu, I. O.; Biciusca, V.; Taisescu, C. I.; Alexandru, D. O.; Taisescu, O.; Comanescu, M. V.; Petrescu, F.; Popescu, I. A.; Trasca, D. M.; ForTofoiu, M. C.; Silosi, C. A.; ForTofoiu, M. Histological factors that predict the liver fibrosis in patients with chronic hepatitis C. Rom. J. Morphol. Embryol. 2016, 57, 759-765.