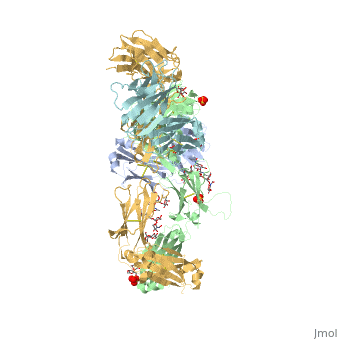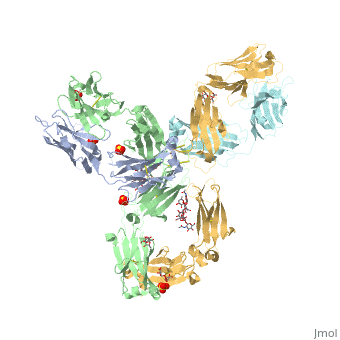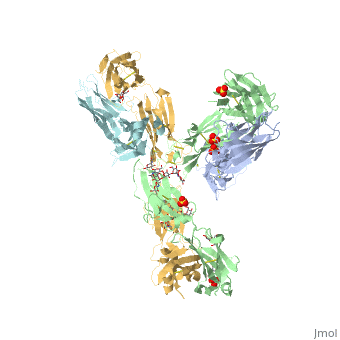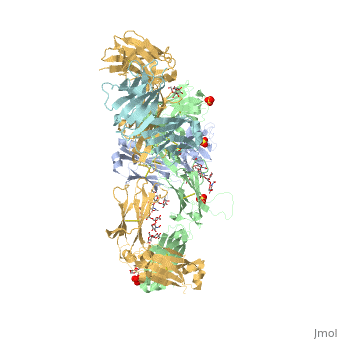We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Keytruda
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==Pembrolizumab antibody against programmed cell death-1 receptor== | ==Pembrolizumab antibody against programmed cell death-1 receptor== | ||
| - | <StructureSection load='5dk3' size='350' side='right' caption='Full-Length Crystal Structure of Pembrolizumab (PDB code [[5dk3]])'> | + | <StructureSection load='5dk3' size='350' side='right' caption='Full-Length Crystal Structure of glycosylated Pembrolizumab complex with sulfate (PDB code [[5dk3]])'> |
== Structure and Function == | == Structure and Function == | ||
| - | Pembrolizumab, trade name Keytruda, is an immunoglobulin G4 (IgG4)-kappa humanized monoclonal antibody against the programmed cell death-1 (PD-1) receptor. It contains an Fv fragment (PemFv) that is the variable region of the molecule where binding | + | '''Pembrolizumab''', trade name '''Keytruda''', is an immunoglobulin G4 (IgG4)-kappa humanized monoclonal antibody against the programmed cell death-1 (PD-1) receptor. It contains an Fv fragment (PemFv) that is the variable region of the molecule where binding occurs, as well as a Fab fragment (PemFab) that constitutes the entire molecule. Pembrolizumab is a very compact molecule with an asymmetrical Y-shape. The short compact hinge region inflicts constraints on the molecule that creates the abnormal crystallizable heavy chain/tail region (Fc domain) compared to other immunoglobulin G (IgG) proteins. The heavy chain is <scene name='74/745945/Glycosylation/1'>glycosylated at Asp297</scene> at both CH<sub>2</sub> domains on each chain and one of them is distinctively rotated 120° compared to other similar structures, making the glycan chain more solvent accessible. IgG4s have a unique function where they form dynamic bispecific antibodies by exchanging half-molecules (one heavy chain/light chain pair) among themselves, called Fab-arm exchange. This makes the molecule particularly unstable and unpredictable as a treatment, but is conquered by introducing the serine-to-proline mutation at <scene name='74/745945/Pro228/1'>amino acid 228</scene>, which prevents Fab-arm exchange and stabilizes the molecule <ref name="log">DOI:10.1080/17425255.2016.1216976</ref>. |
== Mechanism == | == Mechanism == | ||
| + | |||
===Pembrolizumab/PD-1 Interaction=== | ===Pembrolizumab/PD-1 Interaction=== | ||
| - | In order for Pembrolizumab to block PD-1, Pembrolizumab forms a large, flat paratope (antigen-binding site) that can sustain PD-1’s large epitope (where antibody attaches on antigen). The induced interaction between Pembrolizumab and PD-1 gives rise to a surface conformational change on PD-1. The new structure of PD-1 becomes a very shallow, “crescent”-like shape, in contrast to the flat conformation when bound to PD-L1 <ref name="horita">DOI:10.1038/srep35297</ref>. | + | In order for Pembrolizumab to block PD-1, Pembrolizumab forms a large, flat paratope (antigen-binding site) that can sustain PD-1’s large epitope (where antibody attaches on antigen). The induced [http://www.nature.com/articles/srep35297/figures/2 interaction between Pembrolizumab and PD-1] gives rise to a surface conformational change on PD-1. The new structure of PD-1 becomes a very shallow, “crescent”-like shape, in contrast to the flat conformation when bound to PD-L1 <ref name="horita">DOI:10.1038/srep35297</ref>. |
===PemFv/PD-1 Interaction=== | ===PemFv/PD-1 Interaction=== | ||
| - | The Fv fragment of Pembrolizumab can form a complex with the extracellular domain (ECD) of PD-1. Both PemFv and PD-1<sub>ECD</sub> contain interchain disulfide bonds. PemFv interacts predominantly in the major groove of PD-1, which is formed on one surface by the CC’FG antiparallel β−sheet and the BC, C’D, and FG loops. There are 15 direct hydrogen bonds between the residues, 15 water-mediated hydrogen bonds, 2 salt bridges, and many hydrophobic interactions. There are a total of 26 PD-1<sub>ECD</sub> residues involved in the interaction with PemFv, with residues in loop C’D (Pro84 to Gly90) and strand C’ (Gln75 to Lys 78) playing a major role. These key components of PD-1 mainly form interactions through salt bridges and hydrogen bonds with complementary determining regions, the variable domains, of Pembrolizumab. <scene name='74/745945/Chain_b_amino_acids/1'>Thr30, Tyr33, Ser54, | + | The Fv fragment of Pembrolizumab can form a complex with the extracellular domain (ECD) of PD-1. Both PemFv and PD-1<sub>ECD</sub> contain interchain disulfide bonds. PemFv interacts predominantly in the major groove of PD-1, which is formed on one surface by the CC’FG antiparallel β−sheet and the BC, C’D, and FG loops. There are 15 direct hydrogen bonds between the residues, 15 water-mediated hydrogen bonds, 2 salt bridges, and many hydrophobic interactions. There are a total of 26 PD-1<sub>ECD</sub> residues involved in the interaction with PemFv, with residues in loop C’D (Pro84 to Gly90) and strand C’ (Gln75 to Lys 78) playing a major role. These key components of PD-1 mainly form interactions through salt bridges and hydrogen bonds with complementary determining regions, the variable domains, of Pembrolizumab. <scene name='74/745945/Chain_b_amino_acids/1'>Thr30, Tyr33, Ser54, Tyr101, Arg102</scene> on Chain B of Pembrolizumab form bonds with Asp77, Gln75, Lys78, Thr76, Tyr68, and Asn66 of PD-1. It is believed that the sugar chains of PD-1 have no physical contact with Pembrolizumab due to the N-linked glycosylated residues (Asn49, Asn58, Asn74, and Asn116) being located away from the interface <ref name="horita" />. |
===PD-L1/PD-1 Interaction=== | ===PD-L1/PD-1 Interaction=== | ||
| - | The complex formed when protein-derived ligand, PD-L1, interacts with the inhibitory receptor, PD-1, suppresses immune responses against autoantigens and helps in peripheral immune tolerance. However, when tumors over express PD-L1, the interaction with PD-1 inhibits T-lymphocyte proliferation, release of cytokines, and cytotoxicity, exhausting tumor-specific T-cells. There are a total of 12 PD-1<sub>ECD</sub> residues that are involved in forming the complex with the N-terminus of PD-L1<sub>ECD</sub> (PD-L1<sub>ECD-N</sub>). Nine hydrogen bonds, 3 water-mediated hydrogen bonds, 2 salt bridges, and numerous hydrophobic interactions make up the PD-1<sub>ECD</sub>/PD-L1<sub>ECD-N</sub> interaction. The CC’FG sheet within both proteins is the main interaction point. A hydrophobic surface patch is formed when the PD-1<sub>ECD</sub> is in complex with PD-L1<sub>ECD-N</sub>. The PD-1<sub>ECD</sub> residues involved include Val64, Tyr68, Ile126, Leu128, Ala132 and Ile134. Numerous [http://www.nature.com/articles/srep35297/figures/1 Hydrophilic amino acids] that encircle PD-L1<sub>ECD-N</sub> form salt bridges and hydrogen bonds with Asn66, Tyr68, Gln75, Thr76, Asp77, Lys78, Ala132 and Glu136 of PD-1<sub>ECD</sub> <ref name="horita" />. | + | The complex formed when protein-derived ligand, PD-L1, interacts with the inhibitory receptor, PD-1, suppresses immune responses against autoantigens and helps in peripheral immune tolerance. However, when tumors over express PD-L1, the interaction with PD-1 inhibits T-lymphocyte proliferation, release of cytokines, and cytotoxicity, exhausting tumor-specific T-cells. There are a total of 12 PD-1<sub>ECD</sub> residues that are involved in forming the complex with the N-terminus of PD-L1<sub>ECD</sub> (PD-L1<sub>ECD-N</sub>). Nine hydrogen bonds, 3 water-mediated hydrogen bonds, 2 salt bridges, and numerous hydrophobic interactions make up the PD-1<sub>ECD</sub>/PD-L1<sub>ECD-N</sub> interaction. The CC’FG sheet within both proteins is the main interaction point. A hydrophobic surface patch is formed when the PD-1<sub>ECD</sub> is in complex with PD-L1<sub>ECD-N</sub>. The PD-1<sub>ECD</sub> residues involved in this include Val64, Tyr68, Ile126, Leu128, Ala132 and Ile134. Numerous [http://www.nature.com/articles/srep35297/figures/1 Hydrophilic amino acids] that encircle PD-L1<sub>ECD-N</sub> form salt bridges and hydrogen bonds with Asn66, Tyr68, Gln75, Thr76, Asp77, Lys78, Ala132 and Glu136 of PD-1<sub>ECD</sub> <ref name="horita" />. |
== Disease in Humans == | == Disease in Humans == | ||
| Line 20: | Line 21: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | Melanie Kusakavitch also contributed equally to this project, but the computer program is not showing her name due to malfunction. | ||
Current revision
Pembrolizumab antibody against programmed cell death-1 receptor
| |||||||||||
References
- ↑ 1.0 1.1 Longoria TC, Tewari KS. Evaluation of the pharmacokinetics and metabolism of pembrolizumab in the treatment of melanoma. Expert Opin Drug Metab Toxicol. 2016 Oct;12(10):1247-53. doi:, 10.1080/17425255.2016.1216976. Epub 2016 Aug 16. PMID:27485741 doi:http://dx.doi.org/10.1080/17425255.2016.1216976
- ↑ 2.0 2.1 2.2 2.3 Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297
- ↑ Rajakulendran T, Adam DN. Spotlight on pembrolizumab in the treatment of advanced melanoma. Drug Des Devel Ther. 2015 Jun 4;9:2883-6. doi: 10.2147/DDDT.S78036. eCollection, 2015. PMID:26082618 doi:http://dx.doi.org/10.2147/DDDT.S78036
- ↑ Deeks ED. Pembrolizumab: A Review in Advanced Melanoma. Drugs. 2016 Mar;76(3):375-86. doi: 10.1007/s40265-016-0543-x. PMID:26846323 doi:http://dx.doi.org/10.1007/s40265-016-0543-x
Melanie Kusakavitch also contributed equally to this project, but the computer program is not showing her name due to malfunction.