SAICAR synthetase
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load='4fe2' size='340' side='right' caption='Caption for this structure' scene=''> == Function == '''SAICAR synthetase''' (SAI) or '''phosphoribosylaminoimidazole-succi...) |
|||
| (11 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='350' side='right' caption='SAICAR synthetase complex with aminoimidazole-ribonucleotide, ADP, TRIS, aspartate, acetate, Mg+2 and Cl- ions (PDB code [[4fe2]]) ' scene='74/749400/Cv/1'> | |
| - | <StructureSection load=' | + | |
== Function == | == Function == | ||
'''SAICAR synthetase''' (SAI) or '''phosphoribosylaminoimidazole-succinocarboxamide synthetase''' is part of the purine biosynthesis. SAI catalyzes ATP-dependent ligation of carboxyaminoimidazole ribotide (AICAR) with aspartate<ref>PMID:26072057</ref>. | '''SAICAR synthetase''' (SAI) or '''phosphoribosylaminoimidazole-succinocarboxamide synthetase''' is part of the purine biosynthesis. SAI catalyzes ATP-dependent ligation of carboxyaminoimidazole ribotide (AICAR) with aspartate<ref>PMID:26072057</ref>. | ||
| - | |||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
== Structural highlights == | == Structural highlights == | ||
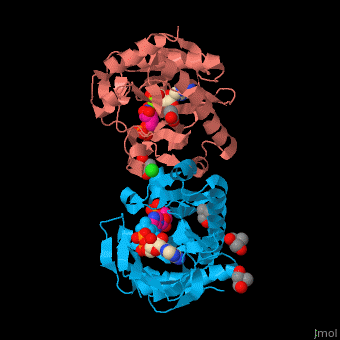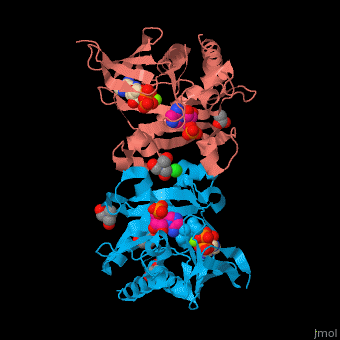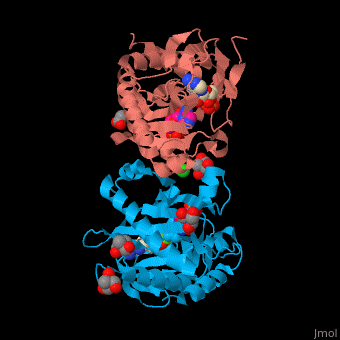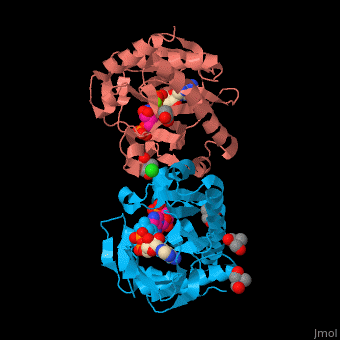
| - | + | SAI structure shows <scene name='74/749400/Cv/10'>two domains</scene>. The <scene name='74/749400/Cv/11'>active site is located in a hydrophilic tunnel</scene> between the 2 domains and contains <scene name='74/749400/Cv/12'>ADP</scene>, <scene name='74/749400/Cv/13'>aspartic acid</scene> and <scene name='74/749400/Cv/14'>aminoimidazole-ribonucleotide (AIR)</scene><ref>PMID:24598753</ref>. Residues are colored according to {{Template:ColorKey_Hydrophobic}}, {{Template:ColorKey_Polar}}. Water molecules are shown as red spheres. | |
| + | *<scene name='74/749400/Cv/15'>Surface representation of hydrophilic tunnel</scene>. | ||
| + | *<scene name='74/749400/Cv/16'>Mg coordination site</scene>. | ||
| + | *<scene name='74/749400/Cv/17'>Whole active site</scene>. | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of | + | ==3D structures of SAICAR synthetase== |
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 20: | Line 18: | ||
**[[1a48]] – ySAI - yeast<br /> | **[[1a48]] – ySAI - yeast<br /> | ||
| - | **[[3r9r]] – | + | **[[3r9r]], [[6yvq]] – MaSAI – ''Mycobacterium abscessus'' <br /> |
| - | **[[3kre]] – SAI – Ehrlichia chaffeensis <br /> | + | **[[3kre]] – SAI – ''Ehrlichia chaffeensis'' <br /> |
| - | **[[1kut]] – SAI – Thermotoga maritima<br /> | + | **[[1kut]] – SAI – ''Thermotoga maritima''<br /> |
*SAICAR synthetase complex | *SAICAR synthetase complex | ||
| - | **[[2yzl]] – MjSAI + ADP – Methanocalcococcus jannaschii <br /> | + | **[[2yzl]] – MjSAI + ADP – ''Methanocalcococcus jannaschii'' <br /> |
**[[2z02]] – MjSAI + ATP <br /> | **[[2z02]] – MjSAI + ATP <br /> | ||
| - | + | **[[4fe2]], [[4nye]] – SpSAI + ADP + aspartate + AICAR – ''Streptococcus pneumoniae''<br /> | |
**[[4fgr]] – SpSAI + ADP <br /> | **[[4fgr]] – SpSAI + ADP <br /> | ||
| - | **[[3nua]] – SAI + ADP + AMP – Clostridium perfringens<br /> | + | **[[3nua]] – SAI + ADP + AMP – ''Clostridium perfringens''<br /> |
| - | **[[2ywv]] – SAI + ADP – Geobsacillus kaustophilus<br /> | + | **[[2ywv]] – SAI + ADP – ''Geobsacillus kaustophilus''<br /> |
**[[2cnq]] – ySAI + ADP + AMP + succinate + AICAR <br /> | **[[2cnq]] – ySAI + ADP + AMP + succinate + AICAR <br /> | ||
**[[2cnv]] – ySAI + aspartate + AICAR derivative<br /> | **[[2cnv]] – ySAI + aspartate + AICAR derivative<br /> | ||
| Line 37: | Line 35: | ||
**[[1obd]] – ySAI + ATP + AMP<br /> | **[[1obd]] – ySAI + ATP + AMP<br /> | ||
**[[1obg]] – ySAI + AMP<br /> | **[[1obg]] – ySAI + AMP<br /> | ||
| - | **[[2gqr]] – EcSAI + ADP – Escherichia coli<br /> | + | **[[2gqr]] – EcSAI + ADP – ''Escherichia coli''<br /> |
**[[2gqs]] – EcSAI + ADP + AICAR <br /> | **[[2gqs]] – EcSAI + ADP + AICAR <br /> | ||
| + | **[[6yy6]], [[6yy7]], [[6yy8]], [[6yy9]], [[6yya]], [[6yyb]], [[6yyc]], [[6yyd]], [[6z0q]], [[6z0r]] – MaSAI + inhibitor<br /> | ||
| + | **[[6yx3]] – MaSAI + ATP <br /> | ||
*SAICAR synthetase type-1 | *SAICAR synthetase type-1 | ||
| - | **[[3u54]] – PhSAI – Pyrococcus horikoshii<br /> | + | **[[3u54]] – PhSAI – ''Pyrococcus horikoshii''<br /> |
**[[4o81]], [[4o82]], [[4o83]] – PhSAI + ADP + AMP <br /> | **[[4o81]], [[4o82]], [[4o83]] – PhSAI + ADP + AMP <br /> | ||
**[[4o84]] – PhSAI + GMP <br /> | **[[4o84]] – PhSAI + GMP <br /> | ||
| Line 61: | Line 61: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of SAICAR synthetase
Updated on 09-March-2022
References
- ↑ Manjunath K, Jeyakanthan J, Sekar K. Catalytic pathway, substrate binding and stability in SAICAR synthetase: A structure and molecular dynamics study. J Struct Biol. 2015 Jul;191(1):22-31. doi: 10.1016/j.jsb.2015.06.006. Epub 2015, Jun 10. PMID:26072057 doi:http://dx.doi.org/10.1016/j.jsb.2015.06.006
- ↑ Wolf NM, Abad-Zapatero C, Johnson ME, Fung LW. Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp. Acta Crystallogr D Biol Crystallogr. 2014 Mar;70(Pt 3):841-50. doi:, 10.1107/S139900471303366X. Epub 2014 Feb 22. PMID:24598753 doi:http://dx.doi.org/10.1107/S139900471303366X