We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Angiotensin-Converting Enzyme
From Proteopedia
(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
{{Clear}} | {{Clear}} | ||
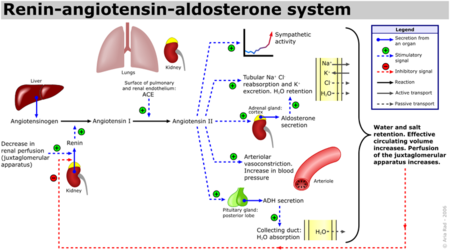
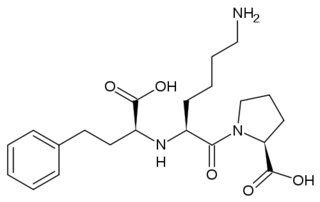
| - | [[Angiotensin-Converting Enzyme]] (ACE) is both an exopeptidase and endopeptindase first discovered by Skeggs et al. in 1956. <ref>Skeggs, L. T., Dorer, F. E., Kahn, J. R., Lentz, K. E., Levin, M. (1981) Experimental renal hypertension: the discovery of the Renin-Angiotensin system. Soffer, R. eds. Biochemical Regulation of Blood Pressure ,3-38 John Wiley & Sons, Inc. Hoboken.</ref> ACE is a zinc- and chloride-dependent metallopeptidase that is responsible for the metabolism of key biologically active peptides, namely Angiotensin I and Bradykinin. These two peptides play a critical role in maintaining appropriate blood pressure in the human body along with a host of other homeostatic circulatory functions. See [[Hypertension & Congestive Heart Failure]]. ACE catalyzes the conversion of the decapeptide | + | [[Angiotensin-Converting Enzyme]] (ACE) is both an exopeptidase and endopeptindase first discovered by Skeggs et al. in 1956. <ref>Skeggs, L. T., Dorer, F. E., Kahn, J. R., Lentz, K. E., Levin, M. (1981) Experimental renal hypertension: the discovery of the Renin-Angiotensin system. Soffer, R. eds. Biochemical Regulation of Blood Pressure ,3-38 John Wiley & Sons, Inc. Hoboken.</ref> ACE is a zinc- and chloride-dependent metallopeptidase that is responsible for the metabolism of key biologically active peptides, namely Angiotensin I and Bradykinin. These two peptides play a critical role in maintaining appropriate blood pressure in the human body along with a host of other homeostatic circulatory functions. See [[Hypertension & Congestive Heart Failure]]. ACE catalyzes the conversion of the decapeptide Angiotensin I to the octapeptide Angiotensin II. Due to its critical role in the Renin-Angiotensin-Aldosterone System (RAAS), ACE has been targeted by a number of pharmaceutical compounds to treat hypertension, diabetic nephropathy, and renal failure. <ref>PMID:10780101</ref> |
<br /> | <br /> | ||
| Line 63: | Line 63: | ||
[[Treatments:ACE Inhibitor Pharmacokinetics References]]<br /> | [[Treatments:ACE Inhibitor Pharmacokinetics References]]<br /> | ||
[[ACE Inhibitor Pharmacokinetics]]<br /> | [[ACE Inhibitor Pharmacokinetics]]<br /> | ||
| + | [[Hypertension & Congestive Heart Failure]]<br /> | ||
| + | [[Lisinopril-Angiotensin Converting Enzyme]]<br /> | ||
| + | [[ACE2 (Hebrew)]]<br /> | ||
[[Treatments:Hypertension]]. | [[Treatments:Hypertension]]. | ||
| - | == | + | ==ACE2 and coronavirus (SARS-CoV and COVID-19) entry into the cell== |
| - | During the SARS scare of 2002-2003, extensive research was focused on the interactions between the SARS virus and its host cells. It was determined that the severe acute respiratory syndrome conavirus (SARS-CoV) enters cells through the activities of a spike shaped protein on its outer envelope. <ref name="SARS">PMID:18448527</ref> The Receptor Binding Domain (RBD) of SARS-CoV binds to ACE2, on the surface of the cell. It was determined that by changing a few selected residues on either the SARS-CoV RBD or the ACE2 binding site, the virus becomes significantly more infectious. <scene name=' | + | During the SARS scare of 2002-2003, extensive research was focused on the interactions between the SARS virus and its host cells. It was determined that the severe acute respiratory syndrome conavirus (SARS-CoV) enters cells through the activities of a spike shaped protein on its outer envelope. <ref name="SARS">PMID:18448527</ref> The Receptor Binding Domain (RBD) of SARS-CoV binds to ACE2, on the surface of the cell. It was determined that by changing a few selected residues on either the SARS-CoV RBD or the ACE2 binding site, the virus becomes significantly more infectious. <scene name='41/413151/Cv/1'>It is believed that these mutations</scene> ([[3d0g]]), namely at residues 31, 35, 38, & 353 in ACE2 or residues 479 and 487 in the SARS-CoV RBD, are what allowed for SARS transmission from [http://en.wikipedia.org/wiki/Civet Civets] to Humans. In fact, in those SARS strains which were determined to be most infectious, the unfavorable electrostatic interactions at the binding interface were removed via mutations at the critical residues 479 and 487. <ref name="SARS"/> |
| - | + | ||
| + | In 2020 Zhou et al. (Nature. 2020; 579: 270-273) and Hoffmann et al. (Cell. 2020; 181: 271-280) showed that SARS-CoV-2, the COVID-19 coronavirus causing the global 2019-2020 pandemia, uses ACE2 as a receptor protein to enter and infect cells, just as SARS-CoV does. Cell entry requires the binding of the S1 region of the virus spike (S) protein to ACE2 followed by the fusion of the viral and cellular membranes produced by the S2 subunit of the S protein. Beforehand, this process requires priming of the S protein by host cell proteases, which is performed by TMPRSS2 and the endosomal cysteine proteases cathepsin B and L (CatB/L). These results suggest therapeutic targets for COVID-19. One is targeting the binding interface between SARS-2-S protein and ACE2, and the other is to inhibit the serine protease activity of the proteases responsible for SARS-2-S protein priming. | ||
==3D Structures of Angiotensin-Converting Enzyme== | ==3D Structures of Angiotensin-Converting Enzyme== | ||
| + | [[Angiotensin-Converting Enzyme 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | *ANCE complexes | ||
| - | **[[2c6n]] - hANCE N domain+lisinopril<br /> | ||
| - | **[[1o86]] - hANCE +lisinopril<br /> | ||
| - | **[[4c2p]] - hANCE + captopril<br /> | ||
| - | **[[2ydm]] - hANCE + captopril analog<br /> | ||
| - | **[[3kbh]], [[3d0g]], [[3d0h]], [[3d0i]], [[2ajf]], [[3sci]], [[3scj]], [[3sck]], [[3scl]] – hANCE 2 fragment+spike glycoprotein<br /> | ||
| - | **[[3l3n]] – hANCE+LISW<br /> | ||
| - | **[[3bkk]], [[3bkl]] – hANCE+ketone inhibitor<br /> | ||
| - | **[[2oc2]], [[4bxk]], [[4ca5]], [[4ca7]], [[4ca8]] - hANCE+phosphinic inhibitor<br /> | ||
| - | **[[2xy9]], [[2xyd]] - hANCE N domain + phosphinic inhibitor<br /> | ||
| - | **[[4ca6]] - hANCE N domain (mutant) + phosphinic inhibitor<br /> | ||
| - | **[[4ufa]] - hANCE N domain (mutant) + acetylserine + aspartate<br /> | ||
| - | **[[4ufb]] - hANCE N domain (mutant) + lysine + proline<br /> | ||
| - | **[[5am8]], [[5am9, [[5ama]], [[5amb]], [[5amc]] - hANCE N domain (mutant) + beta amyloid protein peptide<br /> | ||
| - | **[[1uze]], [[1uzf]] - hANCE+anti-hypertensive drug<br /> | ||
| - | **[[4aph]] – hANCE + angiotensin II<br /> | ||
| - | **[[4apj]] – hANCE + bradykinin-potentiating peptide B<br /> | ||
| - | **[[1r42]], [[1r4l]] – hANCE + disordered segment of collectrin homology domain<br /> | ||
| - | **[[4bzr]] - hANCE+K26<br /> | ||
| - | **[[4bzs]] – hANCE (mutant) +K26<br /> | ||
| - | **[[2xhm]] – DmANCE+K26 <br /> | ||
| - | **[[2x8z]], [[2x90]], [[2x91]], [[2x92]], [[2x93]], [[2x94]], [[2x95]], [[2x96]], [[2x97]], [[1j36]], [[1j37]], [[1j38]] – DmANCE+anti-hypertensive drug<br /> | ||
| - | **[[3zqz]] – DmANCE + captopril analog<br /> | ||
| - | **[[4aa1]] – DmANCE + angiotensin II<br /> | ||
| - | **[[4aa2]], [[4asr]] – DmANCE + bradykinin-potentiating peptide B<br /> | ||
| - | **[[4asq]] – DmANCE + bradykinin peptide | ||
| - | }} | ||
==Additional Resources== | ==Additional Resources== | ||
For Additional Information, see: [[Hypertension & Congestive Heart Failure]] | For Additional Information, see: [[Hypertension & Congestive Heart Failure]] | ||
Current revision
| |||||||||||
Additional Resources
For Additional Information, see: Hypertension & Congestive Heart Failure
References
- ↑ Skeggs, L. T., Dorer, F. E., Kahn, J. R., Lentz, K. E., Levin, M. (1981) Experimental renal hypertension: the discovery of the Renin-Angiotensin system. Soffer, R. eds. Biochemical Regulation of Blood Pressure ,3-38 John Wiley & Sons, Inc. Hoboken.
- ↑ Hoogwerf BJ, Young JB. The HOPE study. Ramipril lowered cardiovascular risk, but vitamin E did not. Cleve Clin J Med. 2000 Apr;67(4):287-93. PMID:10780101
- ↑ 3.0 3.1 3.2 Ferrario CM. Role of angiotensin II in cardiovascular disease therapeutic implications of more than a century of research. J Renin Angiotensin Aldosterone Syst. 2006 Mar;7(1):3-14. PMID:17083068
- ↑ Spyroulias GA, Nikolakopoulou P, Tzakos A, Gerothanassis IP, Magafa V, Manessi-Zoupa E, Cordopatis P. Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship. Eur J Biochem. 2003 May;270(10):2163-73. PMID:12752436
- ↑ 5.0 5.1 Brew K. Structure of human ACE gives new insights into inhibitor binding and design. Trends Pharmacol Sci. 2003 Aug;24(8):391-4. PMID:12915047
- ↑ 6.0 6.1 Sturrock ED, Natesh R, van Rooyen JM, Acharya KR. Structure of angiotensin I-converting enzyme. Cell Mol Life Sci. 2004 Nov;61(21):2677-86. PMID:15549168 doi:10.1007/s00018-004-4239-0
- ↑ 7.0 7.1 7.2 Weir MR. Effects of renin-angiotensin system inhibition on end-organ protection: can we do better? Clin Ther. 2007 Sep;29(9):1803-24. PMID:18035185 doi:10.1016/j.clinthera.2007.09.019
- ↑ Henriksen EJ, Jacob S. Modulation of metabolic control by angiotensin converting enzyme (ACE) inhibition. J Cell Physiol. 2003 Jul;196(1):171-9. PMID:12767053 doi:10.1002/jcp.10294
- ↑ Cole J, Ertoy D, Bernstein KE. Insights derived from ACE knockout mice. J Renin Angiotensin Aldosterone Syst. 2000 Jun;1(2):137-41. PMID:11967804
- ↑ Junot C, Gonzales MF, Ezan E, Cotton J, Vazeux G, Michaud A, Azizi M, Vassiliou S, Yiotakis A, Corvol P, Dive V. RXP 407, a selective inhibitor of the N-domain of angiotensin I-converting enzyme, blocks in vivo the degradation of hemoregulatory peptide acetyl-Ser-Asp-Lys-Pro with no effect on angiotensin I hydrolysis. J Pharmacol Exp Ther. 2001 May;297(2):606-11. PMID:11303049
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 Natesh R, Schwager SL, Sturrock ED, Acharya KR. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003 Jan 30;421(6922):551-4. Epub 2003 Jan 19. PMID:12540854 doi:http://dx.doi.org/10.1038/nature01370
- ↑ Hangauer DG, Monzingo AF, Matthews BW. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochemistry. 1984 Nov 20;23(24):5730-41. PMID:6525336
- ↑ Jaspard E, Alhenc-Gelas F. Catalytic properties of the two active sites of angiotensin I-converting enzyme on the cell surface. Biochem Biophys Res Commun. 1995 Jun 15;211(2):528-34. PMID:7794265
- ↑ http://www.yourlawyer.com/topics/overview/ace_inhibitors
- ↑ Natesh R, Schwager SL, Evans HR, Sturrock ED, Acharya KR. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry. 2004 Jul 13;43(27):8718-24. PMID:15236580 doi:10.1021/bi049480n
- ↑ 16.0 16.1 Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol. 2008 Jul;82(14):6984-91. Epub 2008 Apr 30. PMID:18448527 doi:10.1128/JVI.00442-08
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Cristina Murga