|
|
| (319 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | [[User: Julie Langlois/Sandbox]]
| + | =='''Alginate Binding Periplasmin Proteins of Sphingomonas sp. A1 (AlgQ1, AlgQ2)'''== |
| - | == Overview == | + | <StructureSection load='1y3n' size='350' side='right' caption='Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide (PDB entry [[1y3n]])' scene=''> |
| - | Ribavirin was first synthesized in 1970 by ICN Pharmaceuticals (now “Valent International Pharmaceuticals”). In 1986, its first major use was the treatment of RSV (respiratory syncitial virus) infections in pediatric patients, but since its FDA approval in 1998, it has primarily been used as a component in treating Hepatitis C by inhibiting the synthesis of viral RNA. <ref>https://pubchem.ncbi.nlm.nih.gov/compound/37542<ref> The treatment was modified and approved in 2002 by the FDA by combining it with interferon alfa2b. <ref name="gish"/>
| + | |
| | | | |
| - | == Structure & Function == | |
| | | | |
| | + | == Introduction: == |
| | | | |
| - | While the main function of Ribavirin is to treat Hepatitis C and RSV, Ribavirin alone is not enough to treat these diseases and is commonly combined with interferon alpha2b<ref name="gish"/> . Ribavirin contains antiviral activity which inhibits DNA/RNA synthesis. It is chemically classified as a nucleoside analog because the structure of Ribavirin resembles the structure of the nucleoside guanosine. <ref>https://pubchem.ncbi.nlm.nih.gov/compound/37542<ref>Ribavirin is water soluble and is able to mimic other purines, like guanosin. However, a key difference between the structure of ribavirin and the purine nucleosides is that the heterocyclic base contains only one ring, as opposed to purines which have two. Despite this, it is able to go through similar mechanisms as that of nucleosides, such as phosphorylating into a triphosphate. Its structural similarity to the common nucleoside guanosine may suggest how the drug can inhibit DNA/RNA synthesis through purine mimicry. <ref name="structure"> doi: 10.3851/IMP2125 </ref>
| + | Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer <ref>PMID:15794643</ref>. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1<ref>PMID:8785434</ref>. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M<ref>PMID:22125349</ref>. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa <ref>PMID:15189142</ref>. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter <ref>PMID:9529892</ref>. |
| | + | This structure has <scene name='55/559112/1y3n_ligands/2'>three bound ligands</scene>, <scene name='55/559112/Bem_494_beta-d-mannuronic_acid/1'>BEM</scene>, <scene name='55/559112/Mav_495_alpha-d-mannopyranuron/1'>MAV</scene> and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here.[[image:2dim.jpg|thumb|left|alt=BEM (494) and MAV (495) two-dimensional structures]] |
| | | | |
| - | == Structure & Function == | |
| | | | |
| - | While the main function of Ribavirin is to treat Hepatitis C and RSV, Ribavirin alone is not enough to treat these diseases and is commonly combined with interferon alpha2b<ref name="gish"/> . Ribavirin contains antiviral activity which inhibits DNA/RNA synthesis. It is chemically classified as a nucleoside analog because the structure of Ribavirin resembles the structure of the nucleoside guanosine. <ref>https://pubchem.ncbi.nlm.nih.gov/compound/37542<ref>Ribavirin is water soluble and is able to mimic other purines, like guanosin. However, a key difference between the structure of ribavirin and the purine nucleosides is that the heterocyclic base contains only one ring, as opposed to purines which have two. Despite this, it is able to go through similar mechanisms as that of nucleosides, such as phosphorylating into a triphosphate. Its structural similarity to the common nucleoside guanosine may suggest how the drug can inhibit DNA/RNA synthesis through purine mimicry. <ref name="structure"> doi: 10.3851/IMP2125 </ref>
| + | ---- |
| | | | |
| | + | [[Image:ABC Transporter.jpg|thumb|'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322']] |
| | | | |
| - | == Protein Interaction ==
| + | ---- |
| - | ===Ribavirin with 3SFU===
| + | |
| - | Virally-encoded RNA-dependent RNA polymerase (RdRp) is critical for successful viral replication. The incorporation of nucleoside analogs, such as Ribavirin, by RdRp have been shown to induce error in viral replication
| + | |
| - | Ribavirin attaches to the binding pocket, which is comprised of tyrosine at position 344 and aspartic acid at positions 250, 346, and 347. The incorporation of Ribavirin into the RNA results in base-pairing with uracil or cytosine, which increases the mutation rate and leads to viral death.
| + | |
| - | <scene name='55/559112/Ribavirin_interacting_3sfu/4'>Ribavirin with 3SFU</scene> [[3sfu]]
| + | |
| - | === Ribavirin with 4PB1===
| + | |
| - | Nucleoside transporters facilitate the transport of nucleosides across cell membranes to be used in protein synthesis. Concentrated NTs (CNTs) use the energy of ion gradients for active transportation , while equilibrative NTs (ENTs) transport nucleosides passively down their concentration gradient. Ribavirin is a substrate of ENT1, ENT2, and CNT2. CNT2 binds to the ribose of the ribavirin via hydrogen bonding involving glutamic acid at position 332, asparagine at position 368, and serine at position 371<ref>doi:10.7554/eLife.03604</ref>.
| + | |
| - | <scene name='55/559112/Ribavirin_interacting_4pb1/1'>Ribivirin with 4PB1</scene> [[4pb1]]
| + | |
| | | | |
| | | | |
| | + | == Structural Description and insights to function: == |
| | | | |
| | | | |
| - | == Disease == | + | The alginate binding protein consists of two main groups <scene name='55/559112/Algq1/1'>AlgQ1</scene> and <scene name='55/559112/Algq2/1'>AlgQ2</scene>. AlgQ1 and AlgQ2 are two periplasmic proteins with the almost very similar function which is mediating the transport of the substrate and studies show that the primary structure for this two is about 76% similar, although further investigations are yet to be done to clarify the function and structure differences between these two<ref>PMID:15794643</ref>.Here, we just cover the general structure and functions of these two AlgQ1 and AlgQ2 and their binding to carbohydrates (Alginate). The function of a protein is determined by its shape and the shape of a protein is determined by its primary structure (sequence of amino acids). Based on the analysis of the primary structure done by ([http://www.psort.org/]) and ProtScale ([http://kr.expasy.org/tools/ protscale.html]), this protein is water soluble secretory, which suggests that it is periplasmic<ref>PMID:15794643</ref>. The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria<ref>PMID:29342145</ref>. The N-terminal sequence of AlgQ1 is REATW (Arginine, Glutamic acid, Alanine, Threonine, Tryptophan). The first 24 amino acid residues play the role of a <scene name='55/559112/Signal_peptide/1'>signal peptide</scene>. A signal peptide is a short peptide including between 16 to 30 amino acids, which is found at the N-terminus of proteins that are destined toward the secretory pathway<ref>Kapp, Katja; Schrempf, Sabrina; Lemberg, Marius K.; Dobberstein, Bernhard (2013-01-01). Post-Targeting Functions of Signal Peptides. Landes Bioscience.</ref>. The bulk of this part includes 5 to 16 hydrophobic amino acids. These hydrophobic residues tend to create a single α-helix and are also referred to as “h-region”. there is typically a number of amino acids at the end of the signal peptide that is recognized and cleaved by signal peptidase and therefore named cleavage site<ref>PMID:15794643</ref>. |
| - | ===Hepatitis C===
| + | AlgQ1 in its two forms (Apo and Holo) comprises 490 amino acid residues, indicating that proteins are truncated. The C-terminal amino acid is determined to be Tyrosine. In addition, the amino acid analysis suggests that AlgQ1 is truncated between Tyr490 and Gly491<ref>PMID:15794643</ref>.We can see the apo-AlgQ1 as a cartoon presentation, made up of <scene name='55/559112/Doamins_of_algq1/1'>two globular domains</scene> N domain and c domain, consisting of residues 1-133, 310-400 and 134-309, 401-490 respectively. The two domains that we introduced are connected to each other through 3 loop segments consisting of residues 133-136, 292-347, and 399-401. The disaccharide is bound to the deep cleft between N and C domains. As mentioned in CATH classification, these two domain both are in a class of alpha/beta proteins, showing 3-Layer(aba) Sandwich architecture, having the topology of D-Maltodextrin-Binding Proteins; domain 2 and the homology of both of these domains as can be predicted is Periplasmic binding protein-like II. If we want to assess how SCOP classify this protein, we can see that SCOP sees this protein as a one domain totally, but the same as in CATH, from the alpha and beta proteins. In addition, this protein belongs to Periplasmic binding protein-like superfamily and comes from Phosphate binding protein-like family<ref>PMID:15794643</ref><ref>http://www.cathdb.info/</ref>. |
| - | Hepatitis C is an infectious disease that affects the liver due to the Hepatitis C virus (HCV). This disease can be acute or chronic and can even lead to death. By binding to the gC1qR receptor, HCV proteins are able to effectively inhibit the differentiation of helper T cells. In addition, HCV core proteins work by preventing the synthesis of the antiviral interferon IFN-γ. Thus, weakening the body’s immunity and making it susceptible to infection. Ribavirin is used in combination with peginterferon to treat Hepatitis C.<ref>doi: 10.1002/hep.22070</ref> By adding pegylated interferon-alpha to ribavirin, the drug had a longer half-life, which required only single weekly dosing for Hepatitis C treatment. <ref name="paesh">doi: 10.1016/j.coviro.2011.10.030</ref> There are two types of polyethylene glycol (PEG):1. P-INFa-2b and 2. P-INFa-2a. While P-INFa-2b is linear, covalently attached to histidine, and subject to hydrolysis upon injection, P-INFa-2a is branched, attached to lysine through amide bonds, and circulates as a whole molecule upon injection. The limited distribution of P-INFa-2a results in a longer half-life. <ref>doi:10.2165/11531990-000000000-00000</ref>
| + | When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same<ref>PMID:15794643</ref>. |
| - | ===Pneumonia===
| + | Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively. |
| - | Respiratory syncytial virus is responsible for viral pneumonia. This infection causes the air sacs in one or both of the lungs to become inflamed and potentially filled with fluid or pus. In infants, children, and adults over the age of 65, pneumonia can be deadly. It has been shown that Ribavirin can treat viral pneumonia by preventing transcription of respiratory syncytial virus. <ref> National Heart, Lung and Blood Institute. (2016, September 26). Pneumonia. Retrieved from https://www.nhlbi.nih.gov/health/health-topics/topics/pnu </ref>
| + | The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1<ref>PMID:15794643</ref>. |
| | + | Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)<ref>PMID:15794643</ref>. |
| | | | |
| | + | The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains<ref>PMID:21326935</ref>. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific<ref>PMID:15794643</ref>. |
| | | | |
| - | == Mechanism == | |
| - | Ribavirin, when administered in combination with pegylated interferon alpha, induces an antiviral state in host cells, resulting in reduced virus replication rates and activation of the host immune system. It is thought that Ribavirin acts as an antiviral agent by disrupting viral RNA synthesis, which would impact both transcription and genome replication in the Hepatitis C virus. However, the exact mechanism by which the drug interferes with this process is unknown. Several theories have been proposed to explain the effect of Ribavirin in inhibiting the replication of the Hepatitis C virus <ref name="paesh"/>. | |
| | | | |
| | + | ---- |
| | | | |
| - | Immunomodulation by Ribavirin may be responsible for the drug’s antiviral properties. It has been suggested that the natural CD4+ helper T cell response may be altered in the presence of Ribavirin. It is thought that ribavirin may enhance the T helper 1 response, resulting in greater clearance of virus <ref>doi: 10.1111/j.1478-3231.2008.01896.x</ref>. However, there is conflicting evidence suggesting that the T helper 2 response may be implicated in this process instead <ref>doi: 10.1111/j.1440-1746.2008.05320.x</ref>. Another possible mechanism involves the enhancement of interferon-stimulated gene (ISG) expression by Ribavirin. When a cell becomes infected with a virus, it may release interferons. Interferons are signaling molecules that function in a paracrine fashion to induce an antiviral state in neighboring cells, protecting them from infection. Ribavirin is thought to enhance the interferon signaling pathway <ref>doi: 10.1002/hep.21853</ref>, resulting in a wider antiviral response. This theory has been supported in studies using cell culture models <ref>doi: 10.1002/hep.23985</ref>. It has also been suggested that the relationship between Ribavirin and inosine 5’-monophosphate dehydrogenase (IMPDH) may impact the virus RNA synthesis. IMPDH plays a significant role in the guanine nucleotide synthesis pathway. It results in the conversion of inosine 5’-monophosphate to xanthine 5’-monophosphate, which is an intermediate for the nucleotide guanosine <ref>doi: 10.1002/chin.200823265</ref>. Therefore, modulation of IMPDH activity affects a cell’s reservoir of guanosine. Ribavirin has been shown to function as a competitive inhibitor for IMPDH <ref>PMID: 4197928</ref>. Because guanosine triphosphate (GTP) plays a critical role in the viral genome replication process, inhibition of IMPDH would result in the prevention of viral replication.
| + | ==Links to available structures== |
| | + | Alginate binding periplasmic structures with binding to different alginate polymers |
| | | | |
| | + | [http://www.rcsb.org/structure/1Y3N] :1Y3N |
| | | | |
| - | Ribavirin has also been shown to act as an inhibitor for eIF4E, a protein of the translation initiation complex <ref>doi: 10.1073/pnas.0406927102</ref>. Because of its structural similarity to guanosine, it mimics the 7-methyl guanosine mRNA cap, preventing translation of viral mRNA. This would result in reduced capacity for viral replication within an infected cell. The Hepatitis C viral genome is replicated by RNA-dependent RNA polymerase (RdRp). A modified form of Ribavirin, Ribavirin 5’-triphosphate (RTP) is also believed to directly inhibit RdRp activity <ref>doi: 10.1002/rmv.483</ref>, resulting in lower rates of genome replication. If Ribavirin is converted to the monophosphate form, RMP, it is believed to be incorporated into the viral genome, functioning as a mutagen.
| + | [http://www.rcsb.org/structure/1Y3P] :1Y3P |
| | | | |
| | + | [http://www.rcsb.org/structure/1Y3Q] :1Y3Q |
| | | | |
| | + | [http://www.rcsb.org/structure/1KWH] :1KWH |
| | | | |
| | + | [http://www.rcsb.org/structure/1J1N] :1J1N |
| | | | |
| - | [[Image:Mechanisms.png | thumb]] | + | [http://www.rcsb.org/structure/3A09] :3A09 |
| | | | |
| | + | [http://www.rcsb.org/structure/3VLU] :3VLU |
| | | | |
| | + | [http://www.rcsb.org/structure/3VLV] :3VLV |
| | + | |
| | + | [http://www.rcsb.org/structure/3VLW]: 3VLW |
| | + | |
| | + | [http://www.rcsb.org/structure/5H6U] :5H6U |
| | + | |
| | + | [http://www.rcsb.org/structure/5H71] |
| | + | |
| | + | |
| | + | == Link to evolutionary related Structures== |
| | + | |
| | + | By running the protein through Consurf ([http://consurf.tau.ac.il/2016/]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided. |
| | + | [http://consurf.tau.ac.il//wasabi/?url=http://consurf.tau.ac.il/results/1525350845/query_msa_fasta_and_Tree.xml] |
| | + | |
| | + | The <scene name='55/559112/1y3q_evolutionary/1'>evolutionary conservation 3D structure of 1Y3Q (AlgQ1)</scene>is shown here. |
| | + | |
| | + | |
| | + | ---- |
| | + | |
| | + | == References: == |
| | + | <references /> |
| | </StructureSection> | | </StructureSection> |
| - | == References == | |
| - | # Gish, R. G. Treating HCV with ribavirin analogue and ribavirin-like molecules. Journal of Antimicrobial Chemotherapy. 2005, November 17;1-6. doi:10.1093/jac/dki405 | |
| - | # Thomas, E., Ghany, M., Liang, J., The application and mechanism of action of ribavirin in therapy of hepatitis C. Antiviral Chemistry & Chemotherapy 23:1-12 (2012)doi: 10.3851/IMP2125 | |
| - | # Chung, R.T., Gale, M.J., Polyak, S.J., Lemon, S.M., Liang, T.J., & Hoofnagle, J.H. Mechanisms of action of interferon and ribavirin in chronic hepatitis C: Summary of a workshop. Hepatology. 2008;47 (1), 306-320. doi: 10.1002/hep.22070 | |
| - | # Paeshuyse, J, Dallmeier, K, Neyts, J. Ribavirin for the treatment of chronic hepatitis C virus infection: a review of the proposed mechanisms of action. Current Opinion in Virology. 2011;1(6) 590-598. doi: 10.1016/j.coviro.2011.10.030 | |
| - | # National Heart, Lung and Blood Institute. Pneumonia. 2016, September 26; Retrieved from https://www.nhlbi.nih.gov/health/health-topics/topics/pnu | |
| - | # Foster, G. Pegylated interferons for the treatment of chronic Hepatitis C. Drugs. 2010;70(2):147-165. doi:10.2165/11531990-000000000-00000 | |
| - | #Hofmann WP, Herrmann E, Sarrazin C, Zeuzem S. Ribavirin mode of action in chronic hepatitis C: from clinical use back to molecular mechanisms. Liver Int. 2008, 28:1332-1343. doi: 10.1111/j.1478-3231.2008.01896.x | |
| - | #Alam, I., Lee, J., Cho, K. J., Han, K.R., Yang, J.M., Chung, M.S., & Kim, K. H. Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin. Virology. 2012, 426: 143-151. http://dx.doi.org/10.1016/j.virol.2012.01.016 | |
| - | #Johnson, ZL, Lee, JH, Lee, M, Kwon, DY, Hong, J, Lee, SY. Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters. eLife. 2014, 3:e03604. doi: 10.7554/eLife.03604. | |
| - | #Fujimoto T, Tomimatsu M, Iga D, Endo H, Otsuka K. Changes in the Th1/Th2 ratio during a 24-week course of an interferon alpha-2b plus ribavirin combination therapy for patients with chronic hepatitis C. J. Gastroenterol. Hepatol. 2008, 23:E432- E437. doi: 10.1111/j.1440-1746.2008.05320.x | |
| - | #Feld, JJ, Nanda, S, Huang, Y, Chen, W, Cam, M, Pusek, SN, Schwigler, LM, Theodore, D, Zacks, SL, Liang, TJ, Fried, MW. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecule pathways for treatment response. Hepatol. 2007, 46(5): 1548-1563. doi: 10.1002/hep.21853 | |
| - | #Thomas E, Feld JJ, Li QS, Hu ZY, Fried MW, Liang TJ. Ribavirin potentiates interferon action by augmenting interferon stimulated gene induction in hepatitis C virus cell culture models. Hepatology 2011, 53:32-41. doi: 10.1002/hep.23985 | |
| - | #Shu QN, Nair V. Inosine monophosphate dehydrogenase (IMPDH) as a target in drug discovery. Med. Res. Rev. 2008, 28:219-232. doi: 10.1002/chin.200823265 | |
| - | #Streeter, DG, Witkowski, JT, Khare, GP, Sidwell, RW, Bauer, RJ, Robins, RK, Simon, LN. Mechanisms of action of 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole) a new broad spectrum antiviral agent. Proc. Natl. Acad. Sci. 1973, 70:1174-1178. PMID: 4197928 | |
| - | #Kentsis, A, Topisirovic, I, Culjkovic, B, Shao, L, Borden, KLB. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the guanosine mRNA cap. Proc. Natl. Acad. Sci. 2004, 101:18105-18110. doi: 10.1073/pnas.0406927102 | |
| - | #Graci, JD, Cameron, CE. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006, 16: 37-48. doi: 10.1002/rmv.483 | |
|
Introduction:
Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer [1]. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1[2]. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M[3]. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa [4]. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter [5].
This structure has , , and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here. 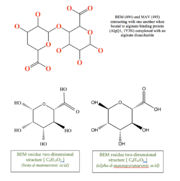 alt=BEM (494) and MAV (495) two-dimensional structures
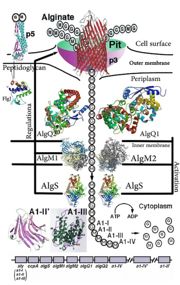 'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi: http://dx.doi.org/10.4161/bbug.1.2.10322'
Structural Description and insights to function:
The alginate binding protein consists of two main groups and . AlgQ1 and AlgQ2 are two periplasmic proteins with the almost very similar function which is mediating the transport of the substrate and studies show that the primary structure for this two is about 76% similar, although further investigations are yet to be done to clarify the function and structure differences between these two[6].Here, we just cover the general structure and functions of these two AlgQ1 and AlgQ2 and their binding to carbohydrates (Alginate). The function of a protein is determined by its shape and the shape of a protein is determined by its primary structure (sequence of amino acids). Based on the analysis of the primary structure done by ([1]) and ProtScale (protscale.html), this protein is water soluble secretory, which suggests that it is periplasmic[7]. The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria[8]. The N-terminal sequence of AlgQ1 is REATW (Arginine, Glutamic acid, Alanine, Threonine, Tryptophan). The first 24 amino acid residues play the role of a . A signal peptide is a short peptide including between 16 to 30 amino acids, which is found at the N-terminus of proteins that are destined toward the secretory pathway[9]. The bulk of this part includes 5 to 16 hydrophobic amino acids. These hydrophobic residues tend to create a single α-helix and are also referred to as “h-region”. there is typically a number of amino acids at the end of the signal peptide that is recognized and cleaved by signal peptidase and therefore named cleavage site[10].
AlgQ1 in its two forms (Apo and Holo) comprises 490 amino acid residues, indicating that proteins are truncated. The C-terminal amino acid is determined to be Tyrosine. In addition, the amino acid analysis suggests that AlgQ1 is truncated between Tyr490 and Gly491[11].We can see the apo-AlgQ1 as a cartoon presentation, made up of N domain and c domain, consisting of residues 1-133, 310-400 and 134-309, 401-490 respectively. The two domains that we introduced are connected to each other through 3 loop segments consisting of residues 133-136, 292-347, and 399-401. The disaccharide is bound to the deep cleft between N and C domains. As mentioned in CATH classification, these two domain both are in a class of alpha/beta proteins, showing 3-Layer(aba) Sandwich architecture, having the topology of D-Maltodextrin-Binding Proteins; domain 2 and the homology of both of these domains as can be predicted is Periplasmic binding protein-like II. If we want to assess how SCOP classify this protein, we can see that SCOP sees this protein as a one domain totally, but the same as in CATH, from the alpha and beta proteins. In addition, this protein belongs to Periplasmic binding protein-like superfamily and comes from Phosphate binding protein-like family[12][13].
When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same[14].
Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively.
The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1[15].
Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)[16].
The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains[17]. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific[18].
Links to available structures
Alginate binding periplasmic structures with binding to different alginate polymers
[2] :1Y3N
[3] :1Y3P
[4] :1Y3Q
[5] :1KWH
[6] :1J1N
[7] :3A09
[8] :3VLU
[9] :3VLV
[10]: 3VLW
[11] :5H6U
[12]
Link to evolutionary related Structures
By running the protein through Consurf ([13]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided.
[14]
The is shown here.
References:
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996 Jun;7(3):301-6. PMID:8785434
- ↑ Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog Polym Sci. 2012 Jan;37(1):106-126. doi: 10.1016/j.progpolymsci.2011.06.003. PMID:22125349 doi:http://dx.doi.org/10.1016/j.progpolymsci.2011.06.003
- ↑ Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241-68. doi: 10.1146/annurev.biochem.73.011303.073626. PMID:15189142 doi:http://dx.doi.org/10.1146/annurev.biochem.73.011303.073626
- ↑ Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998 Mar;62(1):204-29. PMID:9529892
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Miller SI, Salama NR. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018 Jan 17;16(1):e2004935. doi: 10.1371/journal.pbio.2004935., eCollection 2018 Jan. PMID:29342145 doi:http://dx.doi.org/10.1371/journal.pbio.2004935
- ↑ Kapp, Katja; Schrempf, Sabrina; Lemberg, Marius K.; Dobberstein, Bernhard (2013-01-01). Post-Targeting Functions of Signal Peptides. Landes Bioscience.
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ http://www.cathdb.info/
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
|


