User:Pierre Rossignol/Sandbox
From Proteopedia
< User:Pierre Rossignol(Difference between revisions)
| (11 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='4tlf' size='340' side='right' caption='Structure of one subunit of thiol dioxygenase from [https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa Pseudomonas aeruginosa] (PDB code [[4tlf]])' scene=''> | <StructureSection load='4tlf' size='340' side='right' caption='Structure of one subunit of thiol dioxygenase from [https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa Pseudomonas aeruginosa] (PDB code [[4tlf]])' scene=''> | ||
| - | [https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa ''Pseudomonas aeruginosa''] is Gram-negative bacteria. It | + | [https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa ''Pseudomonas aeruginosa''] is classified as a Gram-negative bacteria. It can be described as an opportunistic microorganism since it possesses the ability to colonize bodies of vulnerable patients, especially critically ill ones or those suffering from [https://en.wikipedia.org/wiki/Cystic_fibrosis cystic fibrosis]. ''Pseudomonas aeruginosa'' is one of the major causes of [https://en.wikipedia.org/wiki/Hospital-acquired_infection Hospital-acquired infection] worldwide and a represents a serious threat to Public Health. <ref>https://www.ncbi.nlm.nih.gov/pubmed/21450006</ref> |
| - | ''Pseudomonas aeruginosa'' can colonize many natural environments like soil, water and skin, because of its | + | ''Pseudomonas aeruginosa'' can colonize many natural environments like soil, water and skin, because of its faculty to utilize a wide range of organic matter and cope with different environmental conditions. ''Pseudomonas aeruginosa'' colonizes human hosts, therefore it can use [https://en.wikipedia.org/wiki/Cysteine cysteine] and [https://en.wikipedia.org/wiki/Methionine methionine] as [https://en.wikipedia.org/wiki/Sulfur sulfur] a source of energy. <ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> |
== Function == | == Function == | ||
| - | The [https://en.wikipedia.org/wiki/Thiol thiol] dioxygenation | + | The [https://en.wikipedia.org/wiki/Thiol thiol] dioxygenation represents the initial [https://en.wikipedia.org/wiki/Redox oxidation] step which allows the incorporation of a thiol to catabolic and biosynthetic pathways. A family of specific non-[https://en.wikipedia.org/wiki/Heme heme] mononuclear iron proteins are necessary to catalyse this reaction. During the reaction, each enzyme reacts efficiently with only one substrate. This family of enzymes is made up of cysteine dioxygenase, cysteamine dioxygenase, mercaptosuccinate dioxygenase and 3-mercaptopropionate dioxygenase. <ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> |
| - | The thiol dioxygenase of ''Pseudomonas aeruginosa'' is a 3-mercaptopropionate dioxygenase (p3MDO) with a secondary cysteine dioxygenase activity. Therefore it can also be named 3-mercaptopropionate dioxygenase or cysteine dioxygenase. | + | The thiol dioxygenase of ''Pseudomonas aeruginosa'' is a 3-mercaptopropionate dioxygenase (p3MDO) with a secondary cysteine dioxygenase activity. Therefore, it can also be named 3-mercaptopropionate dioxygenase or cysteine dioxygenase. |
| - | This is the first | + | This is the first example of cysteine dioxygenase homologue which utilizes a second substrate with near stochiometric coupling to dioxygen consumption.<ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> |
The cysteine dioxygenase homologue from ''Pseudomonas aeruginosa'' is expressed in low levels so this metabolic pathway is present in this organism. | The cysteine dioxygenase homologue from ''Pseudomonas aeruginosa'' is expressed in low levels so this metabolic pathway is present in this organism. | ||
| Line 20: | Line 20: | ||
== Interactions: Catalytic activity == | == Interactions: Catalytic activity == | ||
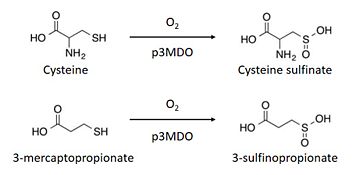
| - | The thiol dioxygenase catalyses the dioxygenation of 3-mercaptopropionate to 3-sulfinopropionate. <ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> | ||
| - | + | [[Image:Thiol_dioxygenase.jpg|thumb|upright=2,5|Thiol dioxygenase of ''Pseudomonas aeruginosa''<ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref>]] | |
| - | 3-mercaptopropionate + O2 = 3-sulfinopropionate | ||
| - | + | The thiol dioxygenase catalyses the dioxygenation of 3-mercaptopropionate to 3-sulfinopropionate. <ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> | |
| - | + | The substrate naturally binds itself to the ferrous iron through the thiol. However, spectroscopy indicates that each substrate can bind themselves to other parts of the enzyme as well. <ref>https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4591825/figure/F1/</ref> | |
| - | + | It also oxidizes cysteine to cysteine sulfinate. <ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> | |
| - | + | This enzyme has a marked preference for 3-mercaptopronionate, which explains why it is also referred to as a 3-mercaptopropionate dioxygenase.<ref>https://www.ncbi.nlm.nih.gov/pubmed/26272617</ref> | |
| Line 38: | Line 36: | ||
== Structural highlights == | == Structural highlights == | ||
| - | Thiol dioxygenases | + | Thiol dioxygenases share a common structure described as a 6-stranded β-barrel core, and a canonical cupin or “jelly roll” β-barrel that is formed with cupin motif 1, an intermotif region, and cupin motif 2 each forming two of the core six β-strands in the folded protein structure.<ref>https://www-ncbi-nlm-nih-gov.scd-rproxy.u-strasbg.fr/pmc/articles/PMC3136866/</ref> |
| - | The Thiol dioxygenase from Pseudomonas aeruginosa is made of 4 chains (named <scene name='75/751223/A/1'>A</scene>, <scene name='75/751223/B/1'>B</scene>, <scene name='75/751223/C/1'>C</scene>, <scene name='75/751223/D/1'>D</scene>)<ref>https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=130072&dps=1</ref>. Each chain is made of 211 amino acids and has a molecular | + | The Thiol dioxygenase from Pseudomonas aeruginosa is made up of 4 chains (named <scene name='75/751223/A/1'>A</scene>, <scene name='75/751223/B/1'>B</scene>, <scene name='75/751223/C/1'>C</scene>, <scene name='75/751223/D/1'>D</scene>)<ref>https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=130072&dps=1</ref>. Each chain is made up of 211 amino acids and has a molecular mass of 23 kDa. For its secondary structures, it has <scene name='75/751223/Alpha_helixes/2'>5 alpha helixes</scene> and <scene name='75/751223/Beta_sheets/1'>14 beta sheets</scene><ref>http://www.uniprot.org/uniprot/Q9I0N5</ref>. Three Histidines, especially,<scene name='75/751223/Histidine_89/1'>Histidine 89</scene>, <scene name='75/751223/His_91/1'>Histidine 91</scene> and <scene name='75/751223/His_142/1'>Histidine 142</scene> constitute the binding sites of iron atom. |
| - | Furthermore, this | + | Furthermore, this protein has two different domains, namely a RmlC-like jelly roll fold and a RmlC-like cupin domain<ref>http://www.ebi.ac.uk/interpro/protein/Q9I0N5</ref>. |
== Disease == | == Disease == | ||
| - | A mammalian cysteine dioxygenase also exists and its active site is slightly different, because of the | + | A mammalian cysteine dioxygenase also exists and its active site is slightly different, because of the presence of a glutamine instead of an arginine. <ref> https://www.ncbi.nlm.nih.gov/pubmed/26272617 </ref> |
In humans, patients with a high level of cysteine and glutathione-cysteine mixed with disulphide are most likely to suffer from [https://en.wikipedia.org/wiki/Pantothenate_kinase-associated_neurodegeneration Hallervorden-Spatz (HS) syndrome] which is essentially characterised by neurochemical abnormalities since it affects the [https://en.wikipedia.org/wiki/Globus_pallidus globus pallidus]. | In humans, patients with a high level of cysteine and glutathione-cysteine mixed with disulphide are most likely to suffer from [https://en.wikipedia.org/wiki/Pantothenate_kinase-associated_neurodegeneration Hallervorden-Spatz (HS) syndrome] which is essentially characterised by neurochemical abnormalities since it affects the [https://en.wikipedia.org/wiki/Globus_pallidus globus pallidus]. | ||
Current revision
Thiol dioxygenase from Pseudomonas aeruginosa
| |||||||||||
References
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/21450006
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4591825/figure/F1/
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www-ncbi-nlm-nih-gov.scd-rproxy.u-strasbg.fr/pmc/articles/PMC3136866/
- ↑ https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=130072&dps=1
- ↑ http://www.uniprot.org/uniprot/Q9I0N5
- ↑ http://www.ebi.ac.uk/interpro/protein/Q9I0N5
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/26272617
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/4073841