This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Epoxidase
From Proteopedia
(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
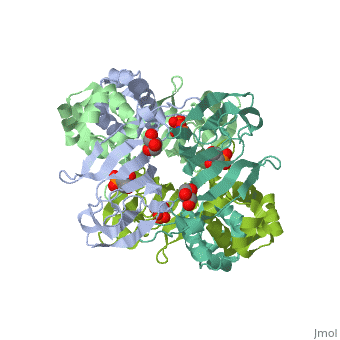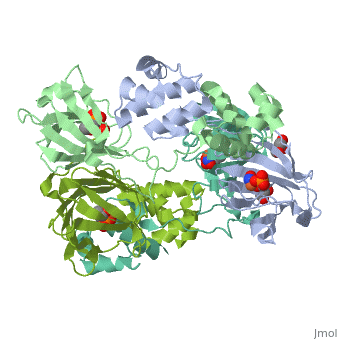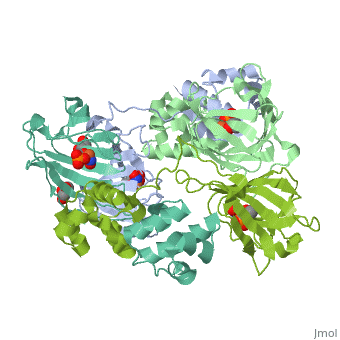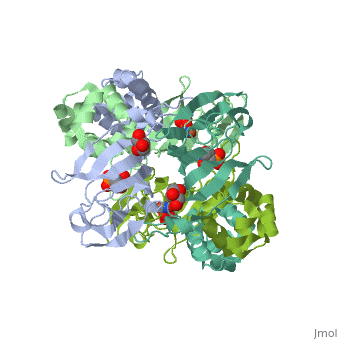
| - | <StructureSection load='3scf' size=' | + | <StructureSection load='3scf' size='350' side='right' caption='Hydroxypropylphosphonic acid epoxidase complex with fosfomycin, glycerol, NO and Fe+2 ion (PDB code [[3scf]])' scene='' pspeed='8'> |
== Function == | == Function == | ||
'''Hydroxypropylphosphonic acid epoxidase''' (HppE) is involved in the biosynthesis of the antibiotic fosfomycin<ref>PMID:16186494</ref>. HppE is a Zn+2/Fe+2-dependent enzyme. '''Halohydrin epoxidase''' (HHE) is involved in the degradation of halohydrins by the displacement of a halogen in halohydrins to produce the epoxide<ref>PMID:26422370</ref>. | '''Hydroxypropylphosphonic acid epoxidase''' (HppE) is involved in the biosynthesis of the antibiotic fosfomycin<ref>PMID:16186494</ref>. HppE is a Zn+2/Fe+2-dependent enzyme. '''Halohydrin epoxidase''' (HHE) is involved in the degradation of halohydrins by the displacement of a halogen in halohydrins to produce the epoxide<ref>PMID:26422370</ref>. | ||
| - | |||
| - | == Disease == | ||
== Relevance == | == Relevance == | ||
| + | Fosfomycin is an antibiotic produced by some ''Streptomyces'' species. | ||
== Structural highlights == | == Structural highlights == | ||
| + | The <scene name='75/752206/Cv/6'>active site of HppE contains fosfomycin and Fe+2</scene>. The enzyme uses an induced-fit mechanism to protect high-energy iron-oxygen species formed during catalysis by moving a <scene name='75/752206/Cv/7'>β-hairpin termed cantilever</scene> to seal off the top portion of the active site<ref>PMID:21682308</ref>. <scene name='75/752206/Cv/8'>Fe coordination site</scene>. | ||
| - | + | ==3D structures of epoxidase== | |
| + | [[Epoxidase 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of epoxidase== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | |||
| - | * 2-hydroxypropylphosphonic acid epoxidase | ||
| - | |||
| - | **[[5u55]], [[5u5d]] – PsPsf4 + Mn + fosfomycin – ''Pseudomonas syringae''<br /> | ||
| - | **[[5u57]], [[5u58]] – PsPsf4 + Fe + fosfomycin <br /> | ||
| - | **[[4j1w]], [[4j1x]], [[3scf]], [[3scg]] – SwHppE + Fe + fosfomycin – ''Strepomyces wedmorensis'' <br /> | ||
| - | **[[1zz6]] – SwHppE <br /> | ||
| - | **[[2bnm]] – SwHppE + Zn <br /> | ||
| - | **[[1zz9]] – SwHppE + Fe <br /> | ||
| - | **[[1zzc]] – SwHppE + Co <br /> | ||
| - | **[[1zz7]], [[1zz8]] – SwHppE + Fe + fosfomycin <br /> | ||
| - | **[[3sch]], [[1zzb]] – SwHppE + Co + fosfomycin <br /> | ||
| - | **[[2bnn]], [[2bno]] – SwHppE + Zn + fosfomycin <br /> | ||
| - | |||
| - | * Halohydrin epoxidase | ||
| - | |||
| - | **[[5b12]] – CoHHE B (mutant) - ''Corynebacterium'' <br /> | ||
| - | **[[4zd9]], [[4zu3]] – CoHHE B <br /> | ||
| - | **[[4z9f]] – CoHHE A (mutant) <br /> | ||
| - | |||
| - | * P450 epoxidase see [[Cytochrome P450]] | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ McLuskey K, Cameron S, Hammerschmidt F, Hunter WN. Structure and reactivity of hydroxypropylphosphonic acid epoxidase in fosfomycin biosynthesis by a cation- and flavin-dependent mechanism. Proc Natl Acad Sci U S A. 2005 Oct 4;102(40):14221-6. Epub 2005 Sep 26. PMID:16186494
- ↑ Watanabe F, Yu F, Ohtaki A, Yamanaka Y, Noguchi K, Yohda M, Odaka M. Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074. Proteins. 2015 Dec;83(12):2230-9. doi: 10.1002/prot.24938. Epub 2015 Oct 16. PMID:26422370 doi:http://dx.doi.org/10.1002/prot.24938
- ↑ Yun D, Dey M, Higgins LJ, Yan F, Liu HW, Drennan CL. Structural Basis of Regiospecificity of a Mononuclear Iron Enzyme in Antibiotic Fosfomycin Biosynthesis. J Am Chem Soc. 2011 Jun 30. PMID:21682308 doi:10.1021/ja2025728