Sandbox Reserved 1056
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_Butler_CH462_Sp2015_#}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_Butler_CH462_Sp2015_#}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
==Carboxypeptidase A in ''B. taurus''== | ==Carboxypeptidase A in ''B. taurus''== | ||
| - | <StructureSection load=' | + | <StructureSection load='1cpx' size='340' side='right' caption='Caption for this structure' scene=''> |
| - | + | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
===Introduction=== | ===Introduction=== | ||
| - | ===Instructions=== | ||
| - | Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
== Biological Function == | == Biological Function == | ||
| Line 16: | Line 12: | ||
== Structural highlights == | == Structural highlights == | ||
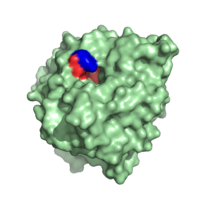
[[Image:Hydrophobic_C-terminal_Side_Chain_Binding_Site_with_Y248.png|200 px|left|thumb|Carboxypeptidase A in ''B. taurus.'' The red highlights the hydrophobic binding pocket while the blue highlights Y248.]] | [[Image:Hydrophobic_C-terminal_Side_Chain_Binding_Site_with_Y248.png|200 px|left|thumb|Carboxypeptidase A in ''B. taurus.'' The red highlights the hydrophobic binding pocket while the blue highlights Y248.]] | ||
| - | + | Shown to the left is the hydrophobic binding pocket of Carboxypeptidase A in ''B. taurus'' in relation to the whole molecule. It is one of the molecule's most important structural features. The <scene name='69/694223/Hydrophobic_binding_pocket_2/2'>hydrophobic binding pocket</scene> is also isolated in the 3D applet to the right. This binding pocket is the site at which C-terminal side chain of the substrate binds. It is formed by amino acid residues I243, I247, A250, G252, G253, S254 and I255. | |
Nearby, there is also Y198 which serves as the recognition site for the sidechain next to the C-terminal residue. | Nearby, there is also Y198 which serves as the recognition site for the sidechain next to the C-terminal residue. | ||
| Line 31: | Line 27: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | |||
| - | Bukrinsky, J.T., Bjerrum, M.J. and Kadziola, A. (1998), Native Carboxypeptidase A in a New Crystal Environment Reveals a Different Conformation of the Important Tyrosine 248. Biochem., 37:16555-16564. | ||
Current revision
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Carboxypeptidase A in B. taurus
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644