User:Loganne Wertz/Sandbox1
From Proteopedia
< User:Loganne Wertz(Difference between revisions)
| (5 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | <StructureSection load='4FXO' size='340' side='right' caption='Caspase-6' scene=''> | ||
| - | Caspase-6 is an [https://en.wikipedia.org/wiki/Endopeptidase endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the [https://en.wikipedia.org/wiki/Caspase cysteine-aspartate family]. Before Caspase-6 becomes functional | + | Caspase-6 is an [https://en.wikipedia.org/wiki/Endopeptidase endopeptidase] involved in apoptosis. In terms of its catalytic function, it is a part of the [https://en.wikipedia.org/wiki/Caspase cysteine-aspartate family]. Before Caspase-6 becomes functional, the enzyme exists as a procaspase, also known as a [https://en.wikipedia.org/wiki/Zymogen zymogen]. In solution, two zymogens are associated together, forming a homodimer. Zymogen activation, the process by which Caspase-6 becomes active, is largely conserved across the caspase family. However, Caspase-6 is unique in that it becomes active through self-cleavage in addition to cleavage by a separate enzyme. Each zymogen of the unprocessed enzyme contains a small subunit consisting of two helices and large subunit consisting of three helices, a prodomain, as well as an intersubunit linker. The helices surround a beta sheet core. In order to become active, the intersubunit linker is bound to the active site of Caspase-6, where it is then cleaved. After cleavage, the four processed subunits, two originating from each zymogen, remain closely associated together through intermolecular forces, forming a dimer of dimers. |
| - | However, Caspase-6 is unique in that it becomes active through self-cleavage | + | |
== Zymogen Activation == | == Zymogen Activation == | ||
| Line 10: | Line 9: | ||
===Active State=== | ===Active State=== | ||
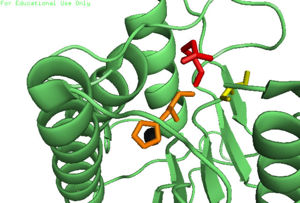
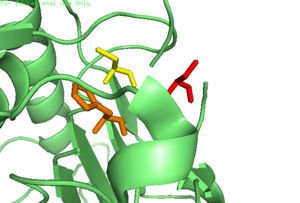
| - | In order to function as an endoprotease, Caspase-6 binds a <scene name='75/752344/Protein_ligand_real/1'>ligand</scene>, which can include neuronal proteins and | + | In order to function as an endoprotease, Caspase-6 binds a <scene name='75/752344/Protein_ligand_real/1'>ligand</scene>, which can include neuronal proteins and [https://en.wikipedia.org/wiki/Tubulin tubulins], in its active site.[[Image:Binding grove active caspase 6.png|100 px|right|thumb|Substrate binding groove in Caspase-6. Blue - catalytic residues |
yellow - ligand | yellow - ligand | ||
red - generic surface]] This binding groove contains three critical amino acid residues necessary to perform cleavage of the peptide bonds. A <scene name='75/752344/Catalytic_triad_real/1'>catalytic triad</scene>[[Image:Cystine Aspartase.png|100 px|right|thumb|active site mechanism]], composed of <scene name='75/752344/His121_real/1'>His-121</scene>, <scene name='75/752344/Glu123_real/1'>Glu-123</scene>, and <scene name='75/752344/Cys163_real/1'>Cys-163</scene>, carries out cleavage of the substrate. In the theorized mechanism, His-121 acts as an acid catalyst, Glu-123 acts as a base catalyst to deprotonate Cys-163, which then acts as covalent catalyst. | red - generic surface]] This binding groove contains three critical amino acid residues necessary to perform cleavage of the peptide bonds. A <scene name='75/752344/Catalytic_triad_real/1'>catalytic triad</scene>[[Image:Cystine Aspartase.png|100 px|right|thumb|active site mechanism]], composed of <scene name='75/752344/His121_real/1'>His-121</scene>, <scene name='75/752344/Glu123_real/1'>Glu-123</scene>, and <scene name='75/752344/Cys163_real/1'>Cys-163</scene>, carries out cleavage of the substrate. In the theorized mechanism, His-121 acts as an acid catalyst, Glu-123 acts as a base catalyst to deprotonate Cys-163, which then acts as covalent catalyst. | ||
==Zinc Inhibition== | ==Zinc Inhibition== | ||
| - | Caspase-6 function is primarily inhibited by the binding of a <scene name='75/752344/Zinc_caspase-6/1'>zinc</scene> ion | + | Caspase-6 function is primarily inhibited by the binding of a <scene name='75/752344/Zinc_caspase-6/1'>zinc</scene> ion, which binds to an <scene name='75/752344/Caspase6_allosteric_site/1'>allosteric site</scene> instead of the <scene name='75/752344/Caspase6_allostericactiv_site/1'>active site</scene>. This allosteric site is located on the outside of the protein and is distal to the active site. The zinc ion is bound to <scene name='75/752344/Caspase6_allosteric_site_resid/1'>three amino acid residues</scene>, Lys-36, Glu-244, and His-287. Once the ion is bound to the protein, it is then stabilized by a <scene name='75/752344/H20_zinc_binding_casp/1'>water molecule</scene> found in the cytoplasm. The binding of zinc at the exosite is suggested to cause a conformational change in the protein from an <scene name='75/752344/Catalytic_triad_real/1'>active state</scene> to an <scene name='75/752344/Inactive_catalytic_triad_casp/1'>inactive state</scene> that misaligns catalytic residues and inhibits activity of the enzyme. It has been proposed that helices of the active dimer must rotate or move in some other way to provide these ideal interactions with zinc. This subtle shift is most likely the cause for allosteric inhibition. As the helices move to bind zinc, the amino acids of the active site become misaligned. The altered positions of the amino acids no longer provide ideal interactions for incoming substrates. After zinc binds, no new substrates enter the active site. Thus, Caspase-6 is effectively inhibited. The residues in the active site no longer provide ideal interactions with the substrate and therefore, substrate does not bind. Zinc binding to the exosite is tightly regulated as it inhibits Caspase-6's critical role in initiation of apoptosis. |
| + | |||
| + | [[Image:4FXO-FINAL.jpg|300px|Caspase-6 w/ Zinc Bound]] [[Image:3s70.jpg|300px|Caspase-6 w/o Zinc Bound]] | ||
===Phosphorylation=== | ===Phosphorylation=== | ||
| Line 22: | Line 23: | ||
=='''Medical Relevance'''== | =='''Medical Relevance'''== | ||
===Caspase-6 involvement in Alzheimer's Disease=== | ===Caspase-6 involvement in Alzheimer's Disease=== | ||
| - | Caspase-6 is known to be involved in many neurodegenerative diseases, one of which is Alzheimer's disease. Caspase-6 activity is associated with the formation of lesions within the | + | Caspase-6 is known to be involved in many neurodegenerative diseases, one of which is Alzheimer's disease (AD). Caspase-6 activity is associated with the formation of lesions within the [http://www.alz.org/ Alzheimer's Disease].Lesions can be found in early stages of AD<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. A proapoptotic protein, p53, is present at increased levels within AD brains, which seems to directly increase the transcription of Caspase-6, which indirectly influences apoptosis of neurons. Future treatments of AD include selective inhibition of active Caspase-6 proteins; staining has found active Caspase-6 within the hippocampus and cortex of the brain within a varying severity of AD cases. This suggests that Caspase-6 plays a predominate role in the pathophysiology of AD. There has been research conducted that shows activation of Caspase-6 in AD could cause disruption of the cytoskeleton network of neurons and lead to neuronal apoptosis<ref name="ActiveRegofCasp6andNDdisease">PMID: 25340928 </ref>. |
Current revision
Caspase-6 in Homo sapiens
| |||||||||||