Structure
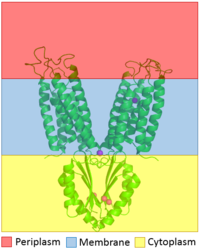
Figure 1. The distribution of YiiP through the membrane is shown. The CTD is shown highlighted in the yellow to show that it is in the cytoplasm and the TMD is highlighted with blue to show that it sits in the membrane
YiiP is a homodimer (protein dimer), with each monomer consisting of 238 residues with TransMembrane () and C-Terminal () domains (colored blue) that are connected via a charge interlocking mechanism () located on a flexible loop. In total, the Yiip protein has three Zn2+ , site A, B, and C. Site A is located in the of the protein, highlighted in purple, site C is located in the , now seen in purple, and site B is located at the junction of the two domains. The TMD, where Zn2+ binding site A resides, consists of 6 transmembrane (TM) helices, 4 of which, (labeled on only one monomer, but present on both), pivot about the ion binding site A. The remaining two helices, , are oriented to the bundle. Movement of these helices plays a role in the function of Zn2+ transport.
A large portion of the protein containing binding site C, the , approximately 30 in length[1], protrudes into the cytoplasm functioning as a Zn2+ sensor within the cell. Zn2+ binding at site C helps hold the CTD together and is thought to stabilize conformational changes in YiiP. YiiP has two different functional conformations which dictates whether or not YiiP is open to the periplasm or the cytoplasm. This salt bridge acts as the hinge for Yiip's conformational changes.
Electrostatic Interactions

Figure 2. Electrostatic Charge Distribution
along the exterior surface of the protein, shown in Figure 1, is primarily neutral (white) for the TMDs, but transitions to positive (blue) near the location of the and interior side of the cell membrane. This positive section is characteristic of trans-membrane proteins as a means of achieving proper orientation within the cell membrane. The A, B, and C, as well as the CTDs of both monomers, all possess a high negative charge (red) relative to the other charges present, facilitating the binding and releasing of Zn
2+ ions. The two CTDs are held together by the charge interlock and hydrophobic interactions of the TMDs, despite their electrostatic repulsion. Upon the release of Zn
2+ ions, the CTDs undergo alterations to electronegativity, which enables domain separation.
Interlocking Salt Bridge
The formation between Lys77 and Asp207 of each domain of YiiP forms an interlocking interaction that acts as the pivot point of the conformational change that drives the function of YiiP as shown in Figure 2. Interlocking interactions are disrupted when Zn
2+ is bound, due to movement of the antiparallel helices, causing a conformational shift in YiiP. The salt bridge also aids in holding the two monomers together, where
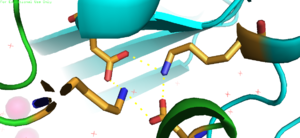
Figure 3. Lys77 and Asp207 Salt Bridges
residues around the salt bridge further stabilize the two domains in the v-shaped void where the domains connect. This prevents degradation of the protein's interlock via interactions with the environment.
Zn2+ Binding Sites
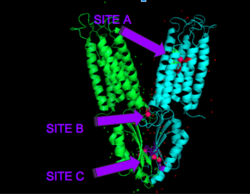
Figure 4. Binding sites A, B, and C
Each Yiip monomer contains three Zn
2+ . There is an active site (Site A), and two cytoplasmic binding sites (Site B and C) depicted in Figure 3. It was found that only site A and C are conserved, while the function of Site B is not well defined, though it is believed that it plays a role in subunit dimerization.
Binding Site A
is in the center of the transmembrane domain, attached and confined via residues from the TM2 and TM5 helices. The TM2 helix has , and the TM5 has , which facilitate the binding and releasing of Zn2+ within the sites. The (shown in blue) is significantly shorter than the other 5 helices around it, and this length forms a cavity in the membrane. Site A is one of Yiip's active sites, where Zn2+ is able to attach and eventually exit the cell via proton transport. This particular site has an ideal tetrahedron among its residues, which is preferred for Zn2+ binding.
It is important to note that the structure of this binding site is rigid because of the coordination of the Zn2+ between the four residues. This rigidity is indicative that any slight shift on either of the helices will cause a drastic readjustment of the coordination of Zn2+. In addition, there are no outer-shell constraints to hold the residues in place, which means that with a readjustment of the molecule, there is no energy being expended to bind or release another Zn2+ molecule. Therefore, the Zn2+ is able to rapidly release and bind a new Zn2+ with a simple reorientation or shift of the molecule.
Binding Site C
has vastly opposite properties from what is seen in binding site A. It is located on a random coil between the two C-terminus domain interfaces. Here, there is a binuclear coordination of Zn2+ between the Asp285 residue that the Zn2+ ions together and the four residues (His232, His248, His283, His161). The Asp285 residue does not have outer shell constraints, however, the same cannot be said for the four histidine residues. These constraints consist of hydrogen bonds to the residues surrounding the binding site allowing bidentate bond formation, or hydrogen bonding to a metal in two places.A couple of these potential hydrogen bonds are shown are shown . Formation of bidentate bonds creates an extensive network of interactions at the CTD interface, which allow for stability and strengthening of the CTD-CTD association.
Mechanism of Transport
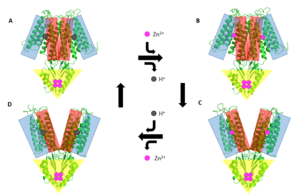
Figure 5. General mechanism that YiiP uses to transport Zn2+ from the cytoplasm to the periplasm. This mechanism involves 2 major conformations; the inward-facing conformation (A & B) and the outward-facing conformation (C & D). Helices TM1, TM2, TM4, and TM5 (blue) are shown pivoting relative to helices TM3 & TM6 (red). The CTD (yellow) does not move during this conformation change as it it held together tightly by binding Zn
2+.
YiiP's ability to export Zn2+ from the cytoplasm is best described as an alternating access mechanism with Zn2+/H+ antiport (Figure 5.). YiiP has 2 major structural conformations as shown by the crystallized structures 3H90 and 3J1Z [1] (a YiiP homolog derived from Shewanella oneidensis). 3H90 shows YiiP in its outward-facing conformation with Zn2+ present and 3J1Z which shows the YiiP homolog in an inward-facing conformation where there is no Zn2+ present.
When YiiP is saturated with Zn2+ it favors the whereas when active sites are either empty or bound to H+ the is favored. This drives the export of Zn2+ from the cytoplasm to the periplasm. Although YiiP exists as a homodimer both monomers can undergo conformation change independent of one other to
produce the alternating access mechanism.
Zn2+ Induced Conformation Change
Zinc induced conformation changes in the TMD and CTD leads to the major and .
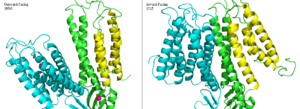
Figure 6. Side by side comparison of one monomer for the the outward-facing conformation of 3H90 and the inward-facing conformation of 3J1Z. TM1, TM2, TM4, and TM5 (yellow) pivot around TM3 and TM6 (green). The helices of the other half of the homodimer (blue) function identically.
The conformation change directly involved with Zn2+/H+ antiport occurs in the TMD as helix pivoting controls what environment site A is available to. Conformation change occurs when the transmembrane helix pairs TM1, TM2, TM4, and TM5 pivot around cation binding site A.[2]
It is believed that the energy for TMD conformation change comes from energy of binding each substrate. Changing to the outward from the inward-facing conformation causes a shift transmembrane helicies TM1, TM2, TM4, and TM5 (Figure 6.) which disrupts Zn2+ binding at site A. Due to the lack of resolution in the 3D structure of 3J1Z a direct comparison of binding site A for each conformation could not be done.[3] This decrease in binding affinity for Zn2+ makes export to the periplasm possible. After Zn2+ is exported and site A is either empty or bound to H+, the protein's conformation changes to the favored inward-facing conformation.
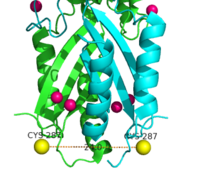
Figure 7. Labeled Cysteine resides measured with FRET showed the distance of the CTD of each monomer to be 24.0Å when saturated with Zn
2+. Decrease in the Cys-Cys distance is indicative that both CTDs of YiiP were brought closer together.
In contrast the main purpose of conformation change in the is to stabilize the YiiP dimer and to act as a Zn2+ sensor.
Using FRET (Figure 7.) to measure the distance between the CTD of each monomer fluorescence quenching was observed as the concentration Zn2+ increased, which supports that idea that Zn2+ induces a stabilizing conformation change in the CTD.[4] CTD of both monomers were measured to be closer together when saturated with Zn2+.
Links to Other YiiP Related Proteopedia Pages
Yiip in action
(3j1z)
(3byp)
Structure of Yiip (2qfi)
References
[5]
[6]
[7]
[8]
[9]
[10]
- ↑ Coudray N, Valvo S, Hu M, Lasala R, Kim C, Vink M, Zhou M, Provasi D, Filizola M, Tao J, Fang J, Penczek PA, Ubarretxena-Belandia I, Stokes DL. Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A. 2013 Jan 22. PMID:23341604 doi:http://dx.doi.org/10.1073/pnas.1215455110
- ↑ Coudray N, Valvo S, Hu M, Lasala R, Kim C, Vink M, Zhou M, Provasi D, Filizola M, Tao J, Fang J, Penczek PA, Ubarretxena-Belandia I, Stokes DL. Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A. 2013 Jan 22. PMID:23341604 doi:http://dx.doi.org/10.1073/pnas.1215455110
- ↑ Coudray N, Valvo S, Hu M, Lasala R, Kim C, Vink M, Zhou M, Provasi D, Filizola M, Tao J, Fang J, Penczek PA, Ubarretxena-Belandia I, Stokes DL. Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A. 2013 Jan 22. PMID:23341604 doi:http://dx.doi.org/10.1073/pnas.1215455110
- ↑ Lu M, Chai J, Fu D. Structural basis for autoregulation of the zinc transporter YiiP. Nat Struct Mol Biol. 2009 Oct;16(10):1063-7. Epub 2009 Sep 13. PMID:19749753 doi:10.1038/nsmb.1662
- ↑ Fu, Min Lu Dax, and Science21 Sep 2007 : 1746-1748. "Structure of the Zinc Transporter YiiP." Structure of the Zinc Transporter YiiP | Science. Science Magazine, n.d. Web. 24 Feb. 2017.
- ↑ "Protein Page: YiiP." Protein Page: YiiP. National Center for Biotechnology Information, n.d. Web. 24 Feb. 2017
- ↑ "Laboratory of David Stokes." NYUSOM. NYU School of Medicine, n.d. Web. 24 Feb. 2017
- ↑ Plum, Laura M., Lothar Rink, and Hajo Haase. "The Essential Toxin: Impact of Zinc on Human Health." International Journal of Environmental Research and Public Health. Molecular Diversity Preservation International (MDPI), Apr. 2010. Web. 24 Feb. 2017.
- ↑ Fu, Dax. Zinc Transporter YiiP Escherichia Coli (n.d.): n. pag. Brookhaven National Laboratory, Mar. 2010. Web. 21 Apr. 2017
- ↑ Paulsen, I.T., and Jr. M.H. Saier. "A Novel Family of Ubiquitous Heavy Metal Ion Transport Proteins." SpringerLink. Springer-Verlag, n.d. Web. 21 Apr. 2017







