DTDP-glucose 4,6-dehydratase
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
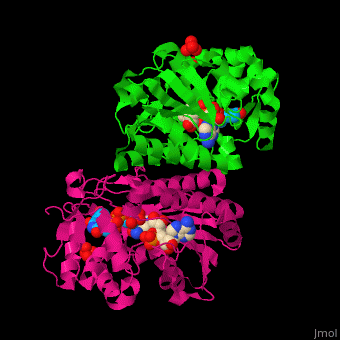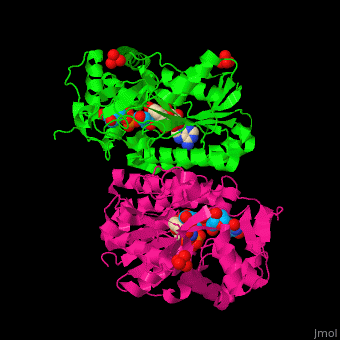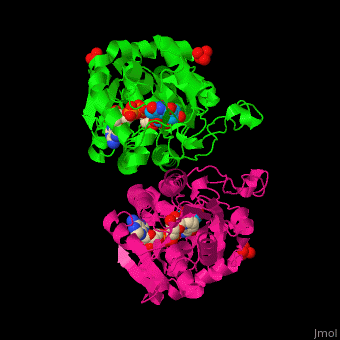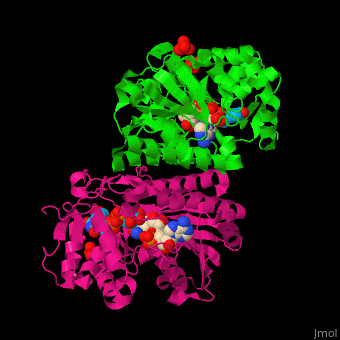
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='RmlB dimer complex with NAD, TDP-D-glucose and sulfate (PDB code [[1ker]])' scene='74/748855/Cv/1'> |
== Function == | == Function == | ||
'''dTDP-glucose 4,6-dehydratase''' (RmlB) catalyzes the reversible conversion of dTDP-glucose to dTDP-4-dehydro-6-deoxy-D-glucose. | '''dTDP-glucose 4,6-dehydratase''' (RmlB) catalyzes the reversible conversion of dTDP-glucose to dTDP-4-dehydro-6-deoxy-D-glucose. | ||
| - | RmlB is the second of four enzymes involved in the dTDP-L-rhamnose pathway<ref>PMID:10666612</ref>. RmlB is involved in the pathway LSP O-antigen biosynthesis which is part of the bacterial outer membrane biogenesis<ref>PMID: | + | RmlB is the second of four enzymes involved in the dTDP-L-rhamnose pathway<ref>PMID:10666612</ref>. RmlB is involved in the pathway LSP O-antigen biosynthesis which is part of the bacterial outer membrane biogenesis<ref>PMID:19019146</ref>. |
== Structural highlights == | == Structural highlights == | ||
The RmlB structure is divided into 2 domains. The N-terminal NAD-binding domain which contains the Rossmann fold and the C-terminal sugar nucleotide binding domain<ref>PMID:11796113</ref>. | The RmlB structure is divided into 2 domains. The N-terminal NAD-binding domain which contains the Rossmann fold and the C-terminal sugar nucleotide binding domain<ref>PMID:11796113</ref>. | ||
| - | *<scene name='74/748855/Cv/ | + | *<scene name='74/748855/Cv/5'>NAD binding site</scene>. |
| - | *<scene name='74/748855/Cv/ | + | *<scene name='74/748855/Cv/6'>TDB-glucose binding site</scene>. |
| - | *<scene name='74/748855/Cv/ | + | *<scene name='74/748855/Cv/7'>Whole active site</scene>. |
</StructureSection> | </StructureSection> | ||
| Line 24: | Line 24: | ||
[[1bxk]] – EcRmlB + NAD – ''Escherichia coli'' <br /> | [[1bxk]] – EcRmlB + NAD – ''Escherichia coli'' <br /> | ||
[[1r66]], [[1r6d]] – RmlB + NAD + TDP derivative – ''Streptococcus venezuelae''<br /> | [[1r66]], [[1r6d]] – RmlB + NAD + TDP derivative – ''Streptococcus venezuelae''<br /> | ||
| - | [[ | + | [[6bi4]] – RmlB + NAD + TDP-sucrose – ''Bacillus anthracis''<br /> |
[[5u4q]] – RmlB + NAD – ''Klebsiella pneumoniae'' <br /> | [[5u4q]] – RmlB + NAD – ''Klebsiella pneumoniae'' <br /> | ||
| + | [[6bwl]] – RmlB + NAD + UDP – ''Bacillus thuringiensis'' <br /> | ||
| + | [[8du1]] – RmlB + NAD – ''Elizabethkingia anophelis'' <br /> | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of RmlB
Updated on 07-February-2023
1kep, 1oc2 – SsRmlB + NAD + TDP-xylose – Streptococcus suis
1ker – SsRmlB + NAD + TDP-glucose
1ket – SsRmlB + NAD + TDP
1keu – StRmlB + NAD + TDP-glucose – Salmonella typhimurium
1kew – StRmlB + NAD + TDP
1g1a – StRmlB + NAD
1bxk – EcRmlB + NAD – Escherichia coli
1r66, 1r6d – RmlB + NAD + TDP derivative – Streptococcus venezuelae
6bi4 – RmlB + NAD + TDP-sucrose – Bacillus anthracis
5u4q – RmlB + NAD – Klebsiella pneumoniae
6bwl – RmlB + NAD + UDP – Bacillus thuringiensis
8du1 – RmlB + NAD – Elizabethkingia anophelis
References
- ↑ Allard ST, Giraud MF, Whitfield C, Messner P, Naismith JH. The purification, crystallization and structural elucidation of dTDP-D-glucose 4,6-dehydratase (RmlB), the second enzyme of the dTDP-L-rhamnose synthesis pathway from Salmonella enterica serovar typhimurium. Acta Crystallogr D Biol Crystallogr. 2000 Feb;56(Pt 2):222-5. PMID:10666612
- ↑ Wang Q, Ding P, Perepelov AV, Xu Y, Wang Y, Knirel YA, Wang L, Feng L. Characterization of the dTDP-D-fucofuranose biosynthetic pathway in Escherichia coli O52. Mol Microbiol. 2008 Dec;70(6):1358-67. doi: 10.1111/j.1365-2958.2008.06449.x., Epub 2008 Oct 30. PMID:19019146 doi:http://dx.doi.org/10.1111/j.1365-2958.2008.06449.x
- ↑ Allard ST, Beis K, Giraud MF, Hegeman AD, Gross JW, Wilmouth RC, Whitfield C, Graninger M, Messner P, Allen AG, Maskell DJ, Naismith JH. Toward a structural understanding of the dehydratase mechanism. Structure. 2002 Jan;10(1):81-92. PMID:11796113