|
|
| (2 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| | | | |
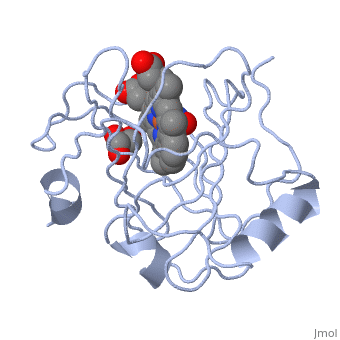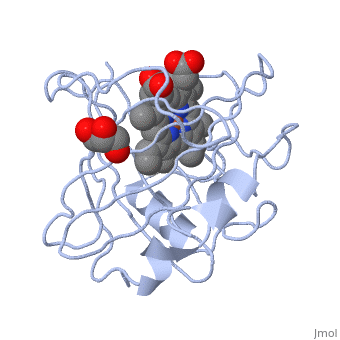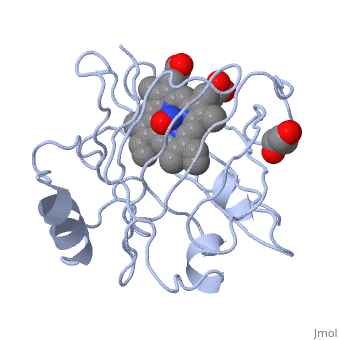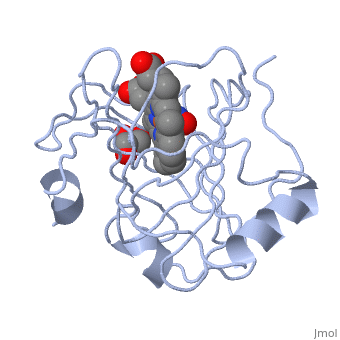
| | ==Crystal structure of nitrophorin 7 from Rhodnius prolixus at pH 7.8 complexed with NO== | | ==Crystal structure of nitrophorin 7 from Rhodnius prolixus at pH 7.8 complexed with NO== |
| - | <StructureSection load='4xme' size='340' side='right' caption='[[4xme]], [[Resolution|resolution]] 1.29Å' scene=''> | + | <StructureSection load='4xme' size='340' side='right'caption='[[4xme]], [[Resolution|resolution]] 1.29Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[4xme]] is a 1 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4XME OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4XME FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[4xme]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Rhodnius_prolixus Rhodnius prolixus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4XME OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4XME FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=NO:NITRIC+OXIDE'>NO</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.29Å</td></tr> |
| - | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Nitrite_dismutase Nitrite dismutase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.7.6.1 1.7.6.1] </span></td></tr>
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=NO:NITRIC+OXIDE'>NO</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4xme FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4xme OCA], [http://pdbe.org/4xme PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=4xme RCSB], [http://www.ebi.ac.uk/pdbsum/4xme PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=4xme ProSAT]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4xme FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4xme OCA], [https://pdbe.org/4xme PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4xme RCSB], [https://www.ebi.ac.uk/pdbsum/4xme PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4xme ProSAT]</span></td></tr> |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[http://www.uniprot.org/uniprot/NP7_RHOPR NP7_RHOPR]] Converts nitrite as the sole substrate to form nitric oxide gas (NO). NO(2-) serves both as an electron donor and as an electron acceptor. Binds to negatively charged cell surfaces of activated platelets; binds to L-a-phosphatidyl-L-serine (PS)-bearing phospholipid membranes. Once bound on an activated platelet, NP7 releases its stored nitric oxide gas (NO) into the victim's tissues while feeding, resulting in vasodilation and inhibition of platelet aggregation. Also acts as an anticoagulant by blocking coagulation-factor binding sites. Has antihistamine activity; binds histamine with high affinity.<ref>PMID:15170336</ref> <ref>PMID:15598503</ref> <ref>PMID:17428677</ref> <ref>PMID:17958381</ref> <ref>PMID:19655755</ref> | + | [https://www.uniprot.org/uniprot/NP7_RHOPR NP7_RHOPR] Converts nitrite as the sole substrate to form nitric oxide gas (NO). NO(2-) serves both as an electron donor and as an electron acceptor. Binds to negatively charged cell surfaces of activated platelets; binds to L-a-phosphatidyl-L-serine (PS)-bearing phospholipid membranes. Once bound on an activated platelet, NP7 releases its stored nitric oxide gas (NO) into the victim's tissues while feeding, resulting in vasodilation and inhibition of platelet aggregation. Also acts as an anticoagulant by blocking coagulation-factor binding sites. Has antihistamine activity; binds histamine with high affinity.<ref>PMID:15170336</ref> <ref>PMID:15598503</ref> <ref>PMID:17428677</ref> <ref>PMID:17958381</ref> <ref>PMID:19655755</ref> |
| | <div style="background-color:#fffaf0;"> | | <div style="background-color:#fffaf0;"> |
| | == Publication Abstract from PubMed == | | == Publication Abstract from PubMed == |
| Line 19: |
Line 19: |
| | </div> | | </div> |
| | <div class="pdbe-citations 4xme" style="background-color:#fffaf0;"></div> | | <div class="pdbe-citations 4xme" style="background-color:#fffaf0;"></div> |
| | + | |
| | + | ==See Also== |
| | + | *[[Nitrophorin|Nitrophorin]] |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Nitrite dismutase]] | + | [[Category: Large Structures]] |
| - | [[Category: Ogata, H]] | + | [[Category: Rhodnius prolixus]] |
| - | [[Category: Heme]] | + | [[Category: Ogata H]] |
| - | [[Category: No transporter]]
| + | |
| - | [[Category: Oxidoreductase]]
| + | |
| - | [[Category: Transport protein]]
| + | |
| Structural highlights
Function
NP7_RHOPR Converts nitrite as the sole substrate to form nitric oxide gas (NO). NO(2-) serves both as an electron donor and as an electron acceptor. Binds to negatively charged cell surfaces of activated platelets; binds to L-a-phosphatidyl-L-serine (PS)-bearing phospholipid membranes. Once bound on an activated platelet, NP7 releases its stored nitric oxide gas (NO) into the victim's tissues while feeding, resulting in vasodilation and inhibition of platelet aggregation. Also acts as an anticoagulant by blocking coagulation-factor binding sites. Has antihistamine activity; binds histamine with high affinity.[1] [2] [3] [4] [5]
Publication Abstract from PubMed
Nitrophorins represent a unique class of heme proteins that are able to perform the delicate transportation and release of the free-radical gaseous messenger nitric oxide (NO) in a pH-triggered manner. Besides its ability to bind to phospholipid membranes, the N-terminus contains an additional Leu-Pro-Gly stretch, which is a unique sequence trait, and the heme cavity is significantly altered with respect to other nitrophorins. These distinctive features encouraged us to solve the X-ray crystallographic structures of NP7 at low and high pH and bound with different heme ligands (nitric oxide, histamine, imidazole). The overall fold of the lipocalin motif is well preserved in the different X-ray structures and resembles the fold of other nitrophorins. However, a chain-like arrangement in the crystal lattice due to a number of head-to-tail electrostatic stabilizing interactions is found in NP7. Furthermore, the X-ray structures also reveal ligand-dependent changes in the orientation of the heme, as well as in specific interactions between the A-B and G-H loops, which are considered to be relevant for the biological function of nitrophorins. Fast and ultrafast laser triggered ligand rebinding experiments demonstrate the pH-dependent ligand migration within the cavities and the exit route. Finally, the topological distribution of pockets located around the heme as well as from inner cavities present at the rear of the protein provides a distinctive feature in NP7, so that while a loop gated exit mechanism to the solvent has been proposed for most nitrophorins, a more complex mechanism that involves several interconnected gas hosting cavities is proposed for NP7.
Structure and dynamics of the membrane attaching nitric oxide transporter nitrophorin 7.,Knipp M, Ogata H, Soavi G, Cerullo G, Allegri A, Abbruzzetti S, Bruno S, Viappiani C, Bidon-Chanal A, Luque FJ F1000Res. 2015 Feb 13;4:45. doi: 10.12688/f1000research.6060.1. eCollection 2015. PMID:26167269[6]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Andersen JF, Gudderra NP, Francischetti IM, Valenzuela JG, Ribeiro JM. Recognition of anionic phospholipid membranes by an antihemostatic protein from a blood-feeding insect. Biochemistry. 2004 Jun 8;43(22):6987-94. PMID:15170336 doi:http://dx.doi.org/10.1021/bi049655t
- ↑ Walker FA. Nitric oxide interaction with insect nitrophorins and thoughts on the electron configuration of the {FeNO}6 complex. J Inorg Biochem. 2005 Jan;99(1):216-36. PMID:15598503 doi:http://dx.doi.org/10.1016/j.jinorgbio.2004.10.009
- ↑ Knipp M, Zhang H, Berry RE, Walker FA. Overexpression in Escherichia coli and functional reconstitution of the liposome binding ferriheme protein nitrophorin 7 from the bloodsucking bug Rhodnius prolixus. Protein Expr Purif. 2007 Jul;54(1):183-91. Epub 2007 Mar 7. PMID:17428677 doi:http://dx.doi.org/10.1016/j.pep.2007.02.017
- ↑ Knipp M, Yang F, Berry RE, Zhang H, Shokhirev MN, Walker FA. Spectroscopic and functional characterization of nitrophorin 7 from the blood-feeding insect Rhodnius prolixus reveals an important role of its isoform-specific N-terminus for proper protein function. Biochemistry. 2007 Nov 20;46(46):13254-68. Epub 2007 Oct 24. PMID:17958381 doi:http://dx.doi.org/10.1021/bi7014986
- ↑ He C, Knipp M. Formation of nitric oxide from nitrite by the ferriheme b protein nitrophorin 7. J Am Chem Soc. 2009 Sep 2;131(34):12042-3. doi: 10.1021/ja9040362. PMID:19655755 doi:http://dx.doi.org/10.1021/ja9040362
- ↑ Knipp M, Ogata H, Soavi G, Cerullo G, Allegri A, Abbruzzetti S, Bruno S, Viappiani C, Bidon-Chanal A, Luque FJ. Structure and dynamics of the membrane attaching nitric oxide transporter nitrophorin 7. F1000Res. 2015 Feb 13;4:45. doi: 10.12688/f1000research.6060.1. eCollection 2015. PMID:26167269 doi:http://dx.doi.org/10.12688/f1000research.6060.1
|