Mur ligase
From Proteopedia
(Difference between revisions)
m (MurD ligase moved to Mur ligase over redirect: requested by Editor) |
|||
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
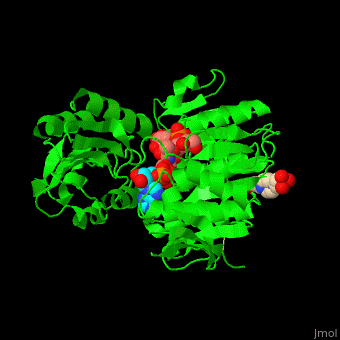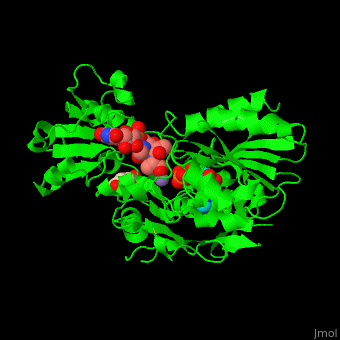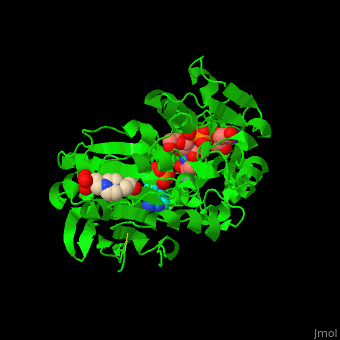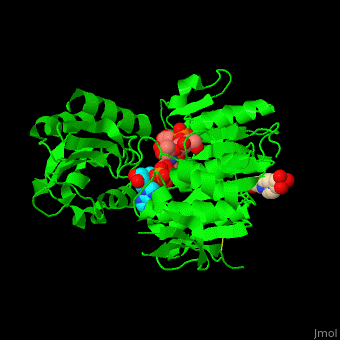
| - | <StructureSection load=' | + | <StructureSection load='' size='350' side='right' caption='Structure of MurD ligase complex with UMA, ADP and Mn+2 ion (purple) (PDB entry [[3uag]])' scene='55/552206/Cv/1'> |
== Function == | == Function == | ||
'''Mur ligase''' are a set of four Mur ubiquitin ligase enzymes: MurC, MurD, MurE, MurF which catalyze the addition of a short polypeptide to UDP-D-acetylmuramic acid in the process of bacterial cell wall buildup from peptidoglycans<ref>PMID:23555903</ref>. All four enzymes are topologically similar. | '''Mur ligase''' are a set of four Mur ubiquitin ligase enzymes: MurC, MurD, MurE, MurF which catalyze the addition of a short polypeptide to UDP-D-acetylmuramic acid in the process of bacterial cell wall buildup from peptidoglycans<ref>PMID:23555903</ref>. All four enzymes are topologically similar. | ||
| Line 12: | Line 12: | ||
== Structural highlights == | == Structural highlights == | ||
| - | <scene name='55/552206/Cv/ | + | <scene name='55/552206/Cv/8'>MurD contains three domains</scene>. |
| - | + | *<scene name='55/552206/Cv/9'>N-terminal domain binds the substrate</scene>. | |
| + | *<scene name='55/552206/Cv/11'>ATP-binding central domain</scene>. C-terminal domain binds the incorporated amino acid<ref>PMID:10356330</ref>. Water molecules are shown as red spheres. | ||
| + | *<scene name='55/552206/Cv/10'>Mn coordination site</scene>, | ||
</StructureSection> | </StructureSection> | ||
| Line 21: | Line 23: | ||
{{#tree:id=OrganizedByTopic|openlevels=0| | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| - | *'''MurD''' | + | *'''MurD''' or '''UDP-N-acetylmuramic acid L-alanyl-D-glutmate ligase''' |
| - | **[[1e0d]] – EcMurD – ''Escherichia coli'' <br /> | + | **[[1e0d]], [[5a5e]] – EcMurD – ''Escherichia coli'' <br /> |
**[[3lk7]] – MurD – ''Streptococcus agalactiae''<br /> | **[[3lk7]] – MurD – ''Streptococcus agalactiae''<br /> | ||
**[[4buc]] – TmMurD – ''Thermotoga maritima''<br /> | **[[4buc]] – TmMurD – ''Thermotoga maritima''<br /> | ||
| Line 33: | Line 35: | ||
**[[2uag]] – EcMurD + UMA + ADP + Mg<br /> | **[[2uag]] – EcMurD + UMA + ADP + Mg<br /> | ||
**[[3uag]] – EcMurD + UMA + ADP + Mn<br /> | **[[3uag]] – EcMurD + UMA + ADP + Mn<br /> | ||
| + | **[[5a5f]] – EcMurD + UMA + ADP + malonate<br /> | ||
**[[4uag]] – EcMurD + UAG<br /> | **[[4uag]] – EcMurD + UAG<br /> | ||
**[[4bub]] – TmMurD + ADP <br /> | **[[4bub]] – TmMurD + ADP <br /> | ||
| Line 42: | Line 45: | ||
**[[2x5o]], [[2y68]], [[2y66]], [[2y67]], [[2y1o]] – EcMurD + thiazolidine inhibitor <br /> | **[[2x5o]], [[2y68]], [[2y66]], [[2y67]], [[2y1o]] – EcMurD + thiazolidine inhibitor <br /> | ||
| - | *MurC | + | *MurC or UDP-N-acetylmuramic acid L-alanine ligase |
**[[1j6u]] – TmMurC <br /> | **[[1j6u]] – TmMurC <br /> | ||
| + | **[[7bva]] – MtMurC – ''Mycobacterium tuberculosis''<br /> | ||
| + | **[[7bvb]] – MtMurC + UAG <br /> | ||
| + | **[[6cau]] – AbMurC + AMPPNP – ''Acinetobacter baumannii'' <br /> | ||
| + | **[[6x9f]], [[6x9n]] – MurC + inhibitor – ''Pseudomonas aeruginosa''<br /> | ||
| + | **[[6vr7]] – MfMurC – ''Methanothermus fervidus'' <br /> | ||
| - | *MurE | + | *MurE or UDP-N-acetylmuramic acid L-alanyl-D-glutmate-2,6-diaminopimelate ligase |
| - | **[[2xja]] – | + | **[[2xja]] – MtMurE + dipeptide + ADP – ''Mycobacterium tuberculosis'' <br /> |
| + | **[[2wtz]] – MtMurE + UAG <br /> | ||
| + | **[[7b53]] – EcMurE <br /> | ||
| + | **[[7b6o]] – EcMurE (mutant)<br /> | ||
| + | **[[1e8c]] – EcMurE + UDP-tripeptide + UAG <br /> | ||
| + | **[[7b60]], [[7b61]], [[7b68]], [[7b6g]], [[7b6i]], [[7b6j]], [[7b6l]], [[7b6k]], [[7b6m]], [[7b6n]], [[7b6q]], [[7b9e]], [[7b9w]] – EcMurE + inhibitor <br /> | ||
| + | **[[7b6p]] – EcMurE (mutant) + inhibitor <br /> | ||
| + | **[[4c12]] – SaMurE + UML + ADP – ''Staphylococcus aureus'' <br /> | ||
| + | **[[4c13]] – SaMurE + UML <br /> | ||
| + | **[[7jt8]] – MfMurE <br /> | ||
| + | **[[6vr8]] – MfMurE + UDP <br /> | ||
| + | **[[7d27]], [[7sir]] – AbMurE <br /> | ||
*MurF | *MurF | ||
| - | **[[2am1]], [[2am2]], [[3zm5]], [[3zm6]] – | + | **[[3ddt]] – hMurF-1 ZF-B box + Zn – human <br /> |
| + | **[[2d8u]] – hMurF-1 ZF-B box + Zn - NMR<br /> | ||
| + | **[[3q1d]] – hMurF-3<br /> | ||
| + | **[[4m3l]] – hMurF-1 coiled coil domain<br /> | ||
| + | **[[2am1]], [[2am2]], [[3zm5]], [[3zm6]] – SpMurF + inhibitor – ''Streptococcus pneumoniae''<br /> | ||
**[[3zl8]] – TmMurF + ADP <br /> | **[[3zl8]] – TmMurF + ADP <br /> | ||
| + | **[[1gg4]] – EcMurF <br /> | ||
| + | **[[4qdi]], [[4qf5]], [[4ziy]] – MurF - ''Acinetobacter baumannii'' <br /> | ||
| + | **[[4cvl]] – PaMurF + AMPPNP - ''Pseudomonas aeruginosa''<br /> | ||
| + | **[[4cvk]] – PaMurF + UDP-MurNac<br /> | ||
| + | **[[4cvm]] – PaMurF + UDP-MurNac + AMPPNP<br /> | ||
| + | *MurT | ||
| + | |||
| + | **[[6fqb]] – SpMurT + glutamine amidotransferase <br /> | ||
| + | **[[6gs2]] – SaMurT + glutamine amidotransferase <br /> | ||
| + | **[[6h5e]] – SaMurT + glutamine amidotransferase + AMPPNP<br /> | ||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D Structures of Mur ligase
Updated on 10-November-2021
References
- ↑ Munshi T, Gupta A, Evangelopoulos D, Guzman JD, Gibbons S, Keep NH, Bhakta S. Characterisation of ATP-dependent Mur ligases involved in the biogenesis of cell wall peptidoglycan in Mycobacterium tuberculosis. PLoS One. 2013;8(3):e60143. doi: 10.1371/journal.pone.0060143. Epub 2013 Mar 21. PMID:23555903 doi:http://dx.doi.org/10.1371/journal.pone.0060143
- ↑ Deva T, Baker EN, Squire CJ, Smith CA. Structure of Escherichia coli UDP-N-acetylmuramoyl:L-alanine ligase (MurC). Acta Crystallogr D Biol Crystallogr. 2006 Dec;62(Pt 12):1466-74. Epub 2006, Nov 23. PMID:17139082 doi:10.1107/S0907444906038376
- ↑ Perdih A, Hodoscek M, Solmajer T. MurD ligase from E. coli: Tetrahedral intermediate formation study by hybrid quantum mechanical/molecular mechanical replica path method. Proteins. 2009 Feb 15;74(3):744-59. doi: 10.1002/prot.22188. PMID:18704940 doi:http://dx.doi.org/10.1002/prot.22188
- ↑ . PMID:212153518
- ↑ Hrast M, Turk S, Sosic I, Knez D, Randall CP, Barreteau H, Contreras-Martel C, Dessen A, O'Neill AJ, Mengin-Lecreulx D, Blanot D, Gobec S. Structure-activity relationships of new cyanothiophene inhibitors of the essential peptidoglycan biosynthesis enzyme MurF. Eur J Med Chem. 2013 May 21;66C:32-45. doi: 10.1016/j.ejmech.2013.05.013. PMID:23786712 doi:10.1016/j.ejmech.2013.05.013
- ↑ Kouidmi I, Levesque RC, Paradis-Bleau C. The biology of Mur ligases as an antibacterial target. Mol Microbiol. 2014 Oct;94(2):242-53. doi: 10.1111/mmi.12758. Epub 2014 Sep 5. PMID:25130693 doi:http://dx.doi.org/10.1111/mmi.12758
- ↑ Bertrand JA, Auger G, Martin L, Fanchon E, Blanot D, Le Beller D, van Heijenoort J, Dideberg O. Determination of the MurD mechanism through crystallographic analysis of enzyme complexes. J Mol Biol. 1999 Jun 11;289(3):579-90. PMID:10356330 doi:10.1006/jmbi.1999.2800
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Jaime Prilusky, Joel L. Sussman