|
|
| (123 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | ==Importance== | + | =='''Alginate Binding Periplasmin Proteins of Sphingomonas sp. A1 (AlgQ1, AlgQ2)'''== |
| - | Lassa virus (LASV), an Old World (OW) [http://en.wikipedia.org/wiki/Arenavirus arenavirus], is a notorious disease-causing agent primarily in West Africa that is able to spread from rodents to humans. This deadly pathogen causes severe viral hemorrhagic fevers and significant mortality. So far, there are no available vaccines for LASV, and only one successful vaccine against another virus found in the ''Arenaviridae'' family: Junin virus<ref name="PMID: 9466512">PMID: 9466512 </ref>. Structural data at atomic resolution for viral proteins are laying the foundation for better understanding both the biology behind viral proteins and ways to combat against them. Determining the structure of the complete trimeric glycoprotein complex (GPC), composed of GP1, GP2, and SSP (stable signal peptide), will pave the path towards a future discovery of novel antiviral drugs. This is the first representative structure for OW arenaviruses. This structure reveals the overall architecture of GP1 domains from OW arenaviruses and important information relating to the mechanisms for pH switching and the binding of LASV to [[LAMP1]] (Lysosome-associated membrane glycoprotein), a recently identified host receptor that is critical for successful infection. In addition, structural analysis suggests two novel immune evasion mechanisms that LASV may utilize to escape antibody-based immune response.
| + | <StructureSection load='1y3n' size='350' side='right' caption='Structure of AlgQ1, alginate-binding protein, complexed with an alginate disaccharide (PDB entry [[1y3n]])' scene=''> |
| - | == Function == | + | |
| - | '''GP1''' (Glycoprotein 1) is the receptor binding domain of LASV that mediates receptor recognition. Research thus far indicates that GP1 from LASV may undergo irreversible conformational changes that could serve as an immunological decoy mechanism. Arenaviruses utilize various cell surface proteins as their cellular receptors for recognizing and attaching to target cells. New World (NW) arenaviruses that belong to clades A and B use transferrin receptor 1 (TfR1)<ref name="PMID:17287727">PMID:17287727</ref><ref name="PMID:24920811">PMID:24920811</ref>, whereas OW arenaviruses, as well as clade C NW arenaviruses, use α-dystroglycan (α-DG) <ref name="PMID:9851928">PMID:9851928</ref><ref name="PMID:15857984">PMID:15857984</ref><ref name="PMID:11967329">PMID:11967329</ref>. A trimeric class 1 viral glycoprotein complex (the spike complex) recognizes the cellular receptors and mediates membrane fusion upon exposure to low pH at the lysosome <ref name="PMID:16731928">PMID:16731928</ref>. The spike complex is expressed as a glycoprotein precursor that is cleaved into three segments by a signal peptidase and SKI-1/S1P protease<ref name="PMID:21612810">PMID:21612810</ref>. The functional spike complex consists of GP1, a membrane-anchored fusion protein (GP2), and a unique structured SSP <ref name="PMID:23202458">PMID:23202458</ref>. | + | |
| - | ==Structural Highlights==
| + | == Introduction: == |
| - | GP1 of LASV is a single chain structure with attached NAG glycans. The overall architecture of GP1 features a central β-sheet and two distinct halves: a glycosylated half containing the receptor-binding site that is made mostly by the central β-sheet and surrounding loops and a half that contains mostly helices and most likely faces the trimer axis)<ref name="PMID: 25972533">PMID: 25972533</ref>. The method used to determine this structure was [http://en.wikipedia.org/wiki/X-ray_crystallography X-ray diffraction]
| + | |
| - | ===LAMP1 Binding Site===
| + | Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer <ref>PMID:15794643</ref>. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1<ref>PMID:8785434</ref>. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M<ref>PMID:22125349</ref>. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa <ref>PMID:15189142</ref>. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter <ref>PMID:9529892</ref>. |
| - | The primary cellular receptor of LASV is α-dystroglycan (α-DG)<ref name="PMID: 9851928">PMID: 9851928</ref><ref name="PMID: 15857984">PMID: 15857984</ref>, which is recognized by a trimeric class 1 viral GPC (spike complex) on the viral surface<ref name="PMID: 16731928">PMID: 16731928</ref><ref name="PMID: 26849049">PMID: 26849049</ref>. Following successful attachment to α-DG on cells, LASV is internalized via [http://en.wikipedia.org/wiki/Pinocytosis macropinocytosis]<ref name="PMID: 27147735">PMID: 27147735</ref>, and the GPC facilitates membrane fusion at the acidic environment of a late endosomal compartment<ref name="PMID: 16731928"><ref name="PMID: 21931550">PMID: 21931550 </ref>. Recent studies have shown that successful infection by LASV requires it to switch in a pH-dependent manner from α-DG to LAMP1<ref name="PMID: 27605678" /><ref name="PMID:24970085">PMID:24970085</ref>. Binding of the LAMP1 endosomal compartment triggers the spikes.
| + | This structure has <scene name='55/559112/1y3n_ligands/2'>three bound ligands</scene>, <scene name='55/559112/Bem_494_beta-d-mannuronic_acid/1'>BEM</scene>, <scene name='55/559112/Mav_495_alpha-d-mannopyranuron/1'>MAV</scene> and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here.[[image:2dim.jpg|thumb|left|alt=BEM (494) and MAV (495) two-dimensional structures]] |
| - | === Histidine Triad=== | + | |
| - | Included in this structure is a unique triad of histidines that is highly conserved among OW arenaviruses. Located on the β-sheet face of GP1, the histidine triad is a structural element that directly interacts with LAMP1 and helps stabilize a LAMP1-"compatible" conformation by providing a molecular mechanism for the pH-dependent receptor switching<ref name="PMID: 25972533" />. The histidine triad is critical in forming a binding site for LAMP1<ref name="PMID: 28448640">PMID: 28448640 </ref><ref name="PMID: 27605678">PMID: 27605678 </ref>.
| + | |
| - | ==Resources== | + | ---- |
| - | For more information on this protein structure visit the following sites: [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4zjf FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4zjf OCA], [http://pdbe.org/4zjf PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=4zjf RCSB], [http://www.ebi.ac.uk/pdbsum/4zjf PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=4zjf ProSAT]
| + | |
| | + | [[Image:ABC Transporter.jpg|thumb|'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322']] |
| | + | |
| | + | ---- |
| | + | |
| | + | |
| | + | == Structural Description and insights to function: == |
| | + | |
| | + | |
| | + | The alginate binding protein consists of two main groups <scene name='55/559112/Algq1/1'>AlgQ1</scene> and <scene name='55/559112/Algq2/1'>AlgQ2</scene>. AlgQ1 and AlgQ2 are two periplasmic proteins with the almost very similar function which is mediating the transport of the substrate and studies show that the primary structure for this two is about 76% similar, although further investigations are yet to be done to clarify the function and structure differences between these two<ref>PMID:15794643</ref>.Here, we just cover the general structure and functions of these two AlgQ1 and AlgQ2 and their binding to carbohydrates (Alginate). The function of a protein is determined by its shape and the shape of a protein is determined by its primary structure (sequence of amino acids). Based on the analysis of the primary structure done by ([http://www.psort.org/]) and ProtScale ([http://kr.expasy.org/tools/ protscale.html]), this protein is water soluble secretory, which suggests that it is periplasmic<ref>PMID:15794643</ref>. The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria<ref>PMID:29342145</ref>. The N-terminal sequence of AlgQ1 is REATW (Arginine, Glutamic acid, Alanine, Threonine, Tryptophan). The first 24 amino acid residues play the role of a <scene name='55/559112/Signal_peptide/1'>signal peptide</scene>. A signal peptide is a short peptide including between 16 to 30 amino acids, which is found at the N-terminus of proteins that are destined toward the secretory pathway<ref>Kapp, Katja; Schrempf, Sabrina; Lemberg, Marius K.; Dobberstein, Bernhard (2013-01-01). Post-Targeting Functions of Signal Peptides. Landes Bioscience.</ref>. The bulk of this part includes 5 to 16 hydrophobic amino acids. These hydrophobic residues tend to create a single α-helix and are also referred to as “h-region”. there is typically a number of amino acids at the end of the signal peptide that is recognized and cleaved by signal peptidase and therefore named cleavage site<ref>PMID:15794643</ref>. |
| | + | AlgQ1 in its two forms (Apo and Holo) comprises 490 amino acid residues, indicating that proteins are truncated. The C-terminal amino acid is determined to be Tyrosine. In addition, the amino acid analysis suggests that AlgQ1 is truncated between Tyr490 and Gly491<ref>PMID:15794643</ref>.We can see the apo-AlgQ1 as a cartoon presentation, made up of <scene name='55/559112/Doamins_of_algq1/1'>two globular domains</scene> N domain and c domain, consisting of residues 1-133, 310-400 and 134-309, 401-490 respectively. The two domains that we introduced are connected to each other through 3 loop segments consisting of residues 133-136, 292-347, and 399-401. The disaccharide is bound to the deep cleft between N and C domains. As mentioned in CATH classification, these two domain both are in a class of alpha/beta proteins, showing 3-Layer(aba) Sandwich architecture, having the topology of D-Maltodextrin-Binding Proteins; domain 2 and the homology of both of these domains as can be predicted is Periplasmic binding protein-like II. If we want to assess how SCOP classify this protein, we can see that SCOP sees this protein as a one domain totally, but the same as in CATH, from the alpha and beta proteins. In addition, this protein belongs to Periplasmic binding protein-like superfamily and comes from Phosphate binding protein-like family<ref>PMID:15794643</ref><ref>http://www.cathdb.info/</ref>. |
| | + | When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same<ref>PMID:15794643</ref>. |
| | + | Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively. |
| | + | The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1<ref>PMID:15794643</ref>. |
| | + | Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)<ref>PMID:15794643</ref>. |
| | + | |
| | + | The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains<ref>PMID:21326935</ref>. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific<ref>PMID:15794643</ref>. |
| | + | |
| | + | |
| | + | ---- |
| | + | |
| | + | ==Links to available structures== |
| | + | Alginate binding periplasmic structures with binding to different alginate polymers |
| | + | |
| | + | [http://www.rcsb.org/structure/1Y3N] :1Y3N |
| | + | |
| | + | [http://www.rcsb.org/structure/1Y3P] :1Y3P |
| | + | |
| | + | [http://www.rcsb.org/structure/1Y3Q] :1Y3Q |
| | + | |
| | + | [http://www.rcsb.org/structure/1KWH] :1KWH |
| | + | |
| | + | [http://www.rcsb.org/structure/1J1N] :1J1N |
| | + | |
| | + | [http://www.rcsb.org/structure/3A09] :3A09 |
| | + | |
| | + | [http://www.rcsb.org/structure/3VLU] :3VLU |
| | + | |
| | + | [http://www.rcsb.org/structure/3VLV] :3VLV |
| | + | |
| | + | [http://www.rcsb.org/structure/3VLW]: 3VLW |
| | + | |
| | + | [http://www.rcsb.org/structure/5H6U] :5H6U |
| | + | |
| | + | [http://www.rcsb.org/structure/5H71] |
| | + | |
| | + | |
| | + | == Link to evolutionary related Structures== |
| | + | |
| | + | By running the protein through Consurf ([http://consurf.tau.ac.il/2016/]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided. |
| | + | [http://consurf.tau.ac.il//wasabi/?url=http://consurf.tau.ac.il/results/1525350845/query_msa_fasta_and_Tree.xml] |
| | + | |
| | + | The <scene name='55/559112/1y3q_evolutionary/1'>evolutionary conservation 3D structure of 1Y3Q (AlgQ1)</scene>is shown here. |
| | + | |
| | + | |
| | + | ---- |
| | + | |
| | + | == References: == |
| | + | <references /> |
| | </StructureSection> | | </StructureSection> |
| - | == References == | |
| - | <references/> | |
| - | [[Category: Cohen, N]] | |
| - | [[Category: Cohen-Dvashi, H]] | |
| - | [[Category: Diskin, R]] | |
| - | [[Category: Israeli, H]] | |
| - | [[Category: Arenavirus]] | |
| - | [[Category: Glycoprotein]] | |
| - | [[Category: Lassa]] | |
| - | [[Category: LASV]] | |
| - | [[Category: Receptor binding]] | |
| - | [[Category: Viral protein]] | |
| - | [[Category: 4zjf]] | |
| - | [[Category: GP1]] | |
|
Introduction:
Alginate binding protein is the protein responsible for mediating the transport of alginate from the pit on the cell surface to the alginate specific ABC (ATP-binding cassette) importer [1]. This system of transferring exists in a gram-negative bacterium, Sphingomonas sp. A1[2]. This protein, which is a periplasmic binding protein, has two homologues AlgQ1 and AlgQ2 coded in PDB as 1Y3N and 1J1N respectively. Alginate is an anionic polysaccharide that is highly found in the cell walls of brown algae. Alginic acid (Alginate) is a linear copolymer with homopolymeric blocks of (1-4)-linked β-D-mannuronate (M) and its C-5 epimer α-L-guluronate (G) residues, linked with covalent bonds. Monomers arrange together in three forms, blocks of consecutive G residues, Blocks of Consecutive M residues and heteropolymeric random sequences of G and M[3]. Strain A1 directly take in this polymeric molecule into the cytoplasm in a process, an important part of which is alginate-binding proteins. 1Y3N is the structure of this alginate binding protein complexed with an alginate disaccharide. A significant number of ABC transporters analyzed so far are just capable of transporting small molecules with a molecular mass less than 2 kDa [4]. In macromolecule assimilation, the macromolecule degrading enzymes play the role of making smaller molecules by breaking macromolecules into pieces, so the living cell can assimilate it. As a result, the alginate ABC importer of strain A1 is unusual in the sense that it can import a macromolecule with an average molecular mass of 26 kDa, without the need to break the macromolecule into smaller parts and the alginate binding protein is the periplasmic binding protein that mediates this transport along with other proteins being active in this specific transporter [5].
This structure has , , and Calcium atom. BEM is beta-D-mannuronic acid, and MAV is alpha-D-mannopyranuronic acid. The two-dimensional structure of these two ligands is provided here. 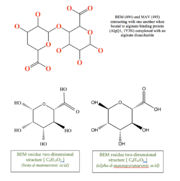 alt=BEM (494) and MAV (495) two-dimensional structures
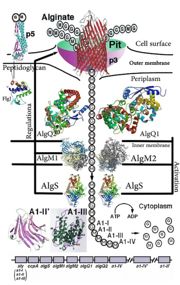 'This is a summary of how Strain A1 superchannel works to import and degrade alginate. G, L-guluronate; M, D-mannuronate; gene for alginate lyases (A1-I, A1-II and A1-III); catabolite-control protein gene; algS, algM1 and algM2, ABC transporter genes for alginate import; algQ1 and algQ2, genes for alginate-binding proteins; a1-IV, alginate lyase A1-IV gene; a1-II’, alginate lyase A1-II’ gene; a1-IV’, alginate lyase A1-IV’ gene; p3, TonB-dependent transporter; p5, alginate receptor; FlgJ, C-terminal catalytic module for peptidoglycan hydrolysis. Adopted from Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi: http://dx.doi.org/10.4161/bbug.1.2.10322'
Structural Description and insights to function:
The alginate binding protein consists of two main groups and . AlgQ1 and AlgQ2 are two periplasmic proteins with the almost very similar function which is mediating the transport of the substrate and studies show that the primary structure for this two is about 76% similar, although further investigations are yet to be done to clarify the function and structure differences between these two[6].Here, we just cover the general structure and functions of these two AlgQ1 and AlgQ2 and their binding to carbohydrates (Alginate). The function of a protein is determined by its shape and the shape of a protein is determined by its primary structure (sequence of amino acids). Based on the analysis of the primary structure done by ([1]) and ProtScale (protscale.html), this protein is water soluble secretory, which suggests that it is periplasmic[7]. The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria[8]. The N-terminal sequence of AlgQ1 is REATW (Arginine, Glutamic acid, Alanine, Threonine, Tryptophan). The first 24 amino acid residues play the role of a . A signal peptide is a short peptide including between 16 to 30 amino acids, which is found at the N-terminus of proteins that are destined toward the secretory pathway[9]. The bulk of this part includes 5 to 16 hydrophobic amino acids. These hydrophobic residues tend to create a single α-helix and are also referred to as “h-region”. there is typically a number of amino acids at the end of the signal peptide that is recognized and cleaved by signal peptidase and therefore named cleavage site[10].
AlgQ1 in its two forms (Apo and Holo) comprises 490 amino acid residues, indicating that proteins are truncated. The C-terminal amino acid is determined to be Tyrosine. In addition, the amino acid analysis suggests that AlgQ1 is truncated between Tyr490 and Gly491[11].We can see the apo-AlgQ1 as a cartoon presentation, made up of N domain and c domain, consisting of residues 1-133, 310-400 and 134-309, 401-490 respectively. The two domains that we introduced are connected to each other through 3 loop segments consisting of residues 133-136, 292-347, and 399-401. The disaccharide is bound to the deep cleft between N and C domains. As mentioned in CATH classification, these two domain both are in a class of alpha/beta proteins, showing 3-Layer(aba) Sandwich architecture, having the topology of D-Maltodextrin-Binding Proteins; domain 2 and the homology of both of these domains as can be predicted is Periplasmic binding protein-like II. If we want to assess how SCOP classify this protein, we can see that SCOP sees this protein as a one domain totally, but the same as in CATH, from the alpha and beta proteins. In addition, this protein belongs to Periplasmic binding protein-like superfamily and comes from Phosphate binding protein-like family[12][13].
When N-domains of apo and holo forms of the protein were superimposed, then a rotation angle of 0.6 degrees was needed for the C domain to be superimposed. Structures of a holo-AlgQ with tetrasaccharide and disaccharide (AlgQ1) are almost the same[14].
Here we can see the binding sites holo-algQ1-DI. This bound consists of ΔM1-M2 with α-anomeric M2 at S1 and S2. ΔM, M, and G denote unsaturated D-mannuronate, saturated D-mannuronate, and saturated L-guluronate, respectively.
The bound oligosaccharides interact with surrounding amino acids. As Keiko momma, et al. summarized hydrogen bond interactions between the bound alginate oligosaccharides and alginate binding proteins(data not shown) The number of direct hydrogen bonds between AlgQ1 and the disaccharide in holo-AlgQ1 DI is 11 and the number of associated water molecules is 10. Five water molecules are located at S3 and S4 subsites in holo-AlgQ1-DI. The number of C-C contacts that AlgQ1 and disaccharide holo-AlgQ1-DI have is 30, which indicates that the nonreducing end of sugar is in a significant involvement with AlgQ1[15].
Ramachandran plot analysis shows that most non-glycine residues are located in the most favorable regions. There is only one exception to this, which is Lys251, (apo-AlgQ1, φ=65° and ψ=-141°; holo-AlgQ1-TE, φ=62° and ψ=-133°; and holo-AlgQ1-DI (1Y3N), φ=65° and ψ=-139°), and it is present in an allowed region. Lys251 is located next to the terminus of a helix (H12/C)[16].
The crystal structure of AlgQ2 consist of two domains separated by a cleft and binds and releases alginate tetrasaccharide by creating conformational change in these two domains[17]. To mention some of the different forms of this protein we can take a look at 5H6U, 5H71, 1KWH, 1J1N in PDB. As an alginate binding protein, the dissociation constants are 6 μg/mL for AlgQ1 and 4 μg/mL for AlgQ2.Results from UV absorption spectroscopy indicate that both of these proteins are alginate specific[18].
Links to available structures
Alginate binding periplasmic structures with binding to different alginate polymers
[2] :1Y3N
[3] :1Y3P
[4] :1Y3Q
[5] :1KWH
[6] :1J1N
[7] :3A09
[8] :3VLU
[9] :3VLV
[10]: 3VLW
[11] :5H6U
[12]
Link to evolutionary related Structures
By running the protein through Consurf ([13]), the Phylogenetic tree of 1Y3N can be obtained. Below a link to this tree is provided.
[14]
The is shown here.
References:
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996 Jun;7(3):301-6. PMID:8785434
- ↑ Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog Polym Sci. 2012 Jan;37(1):106-126. doi: 10.1016/j.progpolymsci.2011.06.003. PMID:22125349 doi:http://dx.doi.org/10.1016/j.progpolymsci.2011.06.003
- ↑ Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241-68. doi: 10.1146/annurev.biochem.73.011303.073626. PMID:15189142 doi:http://dx.doi.org/10.1146/annurev.biochem.73.011303.073626
- ↑ Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998 Mar;62(1):204-29. PMID:9529892
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Miller SI, Salama NR. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018 Jan 17;16(1):e2004935. doi: 10.1371/journal.pbio.2004935., eCollection 2018 Jan. PMID:29342145 doi:http://dx.doi.org/10.1371/journal.pbio.2004935
- ↑ Kapp, Katja; Schrempf, Sabrina; Lemberg, Marius K.; Dobberstein, Bernhard (2013-01-01). Post-Targeting Functions of Signal Peptides. Landes Bioscience.
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ http://www.cathdb.info/
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
- ↑ Hashimoto W, Kawai S, Murata K. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs. 2010 Mar-Apr;1(2):97-109. doi: 10.4161/bbug.1.2.10322. Epub 2009 Oct, 14. PMID:21326935 doi:http://dx.doi.org/10.4161/bbug.1.2.10322
- ↑ Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005 Apr 5;44(13):5053-64. PMID:15794643 doi:10.1021/bi047781r
|


