Phosphoserine phosphatase
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
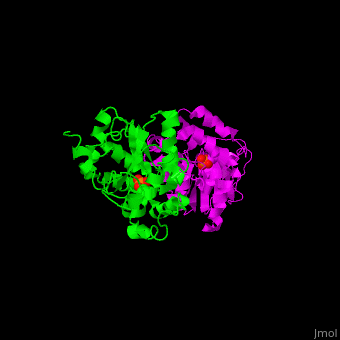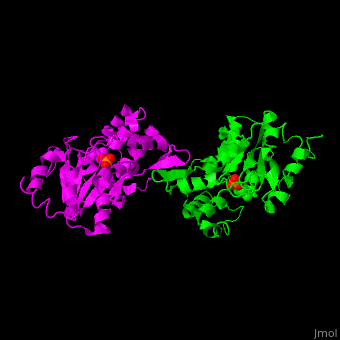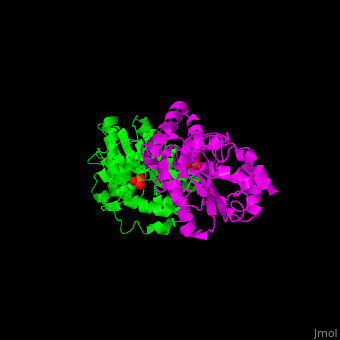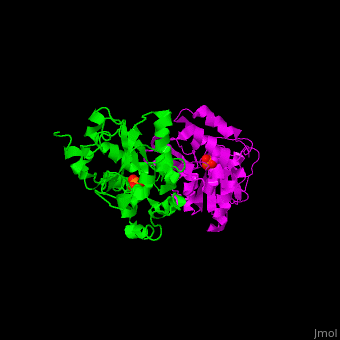
<StructureSection load='' size='450' side='right' scene='48/488517/Cv/1' caption='Human phosphoserine phosphatase dimer complex with serine and phosphate [[1l8o]]'> | <StructureSection load='' size='450' side='right' scene='48/488517/Cv/1' caption='Human phosphoserine phosphatase dimer complex with serine and phosphate [[1l8o]]'> | ||
== Function == | == Function == | ||
| - | '''Phosphoserine phosphatase''' (PSP) catalyzes the dephosphorylation of phosphoserine to produce serine and phosphate. This is the last reaction in serine biosynthesis. PSP is part of the glycine, serine and threonine metabolism<ref>PMID:13610904</ref>. Mg+2 ion is a cofactor of PSP. | + | '''Phosphoserine phosphatase''' (PSP) catalyzes the dephosphorylation of phosphoserine to produce serine and phosphate. This is the last reaction in serine biosynthesis. PSP is part of the glycine, serine and threonine metabolism<ref>PMID:13610904</ref>. Mg<sup>+2</sup> ion is a cofactor of PSP. |
== Disease == | == Disease == | ||
| Line 7: | Line 7: | ||
== Structural highlights == | == Structural highlights == | ||
| - | The human PSP overall structure contains a <scene name='48/488517/Cv/ | + | The human PSP overall structure contains a <scene name='48/488517/Cv/7'>large core domain and a smaller 4-helix bundle domain</scene>. The <scene name='48/488517/Cv/8'>active site</scene> is located in a <scene name='48/488517/Cv/9'>cleft between the 2 domains</scene><ref>PMID:12213811</ref>. |
</StructureSection> | </StructureSection> | ||
==3D structures of phosphoserine phosphatase== | ==3D structures of phosphoserine phosphatase== | ||
| Line 21: | Line 21: | ||
**[[4ij5]] – HtPSP – ''Hydrogenobacter thermophilus''<br /> | **[[4ij5]] – HtPSP – ''Hydrogenobacter thermophilus''<br /> | ||
**[[3n28]] – PSP – ''Vibrio cholerae''<br /> | **[[3n28]] – PSP – ''Vibrio cholerae''<br /> | ||
| + | **[[5zkk]] – EhPSP – ''Entamoeba histolitica''<br /> | ||
*Phosphoserine phosphatase binary complex | *Phosphoserine phosphatase binary complex | ||
| + | **[[1l8l]] – hPSP (mutant) + phosphono-propionic acid – human<br /> | ||
| + | **[[1nnl]], [[6hyj]] – hPSP + Ca<br /> | ||
**[[3p96]], [[5is2]] – MaPSP + Mg – ''Mycobacterium avium''<br /> | **[[3p96]], [[5is2]] – MaPSP + Mg – ''Mycobacterium avium''<br /> | ||
**[[5it0]], [[5it4]], [[5jjb]], [[5t41]] – MaPSP (mutant) + Mg<br /> | **[[5it0]], [[5it4]], [[5jjb]], [[5t41]] – MaPSP (mutant) + Mg<br /> | ||
**[[3m1y]] – PSP + Mg – ''Helicobacter pylori''<br /> | **[[3m1y]] – PSP + Mg – ''Helicobacter pylori''<br /> | ||
**[[1l7o]] – MjPSP (mutant) + Zn – ''Methanocaldococcus jannaschi''i<br /> | **[[1l7o]] – MjPSP (mutant) + Zn – ''Methanocaldococcus jannaschi''i<br /> | ||
| - | **[[1l8l]] – hPSP (mutant) + phosphono-propionic acid – human<br /> | ||
| - | **[[1nnl]] – hPSP + Ca<br /> | ||
**[[3w40]], [[3w41]] – BsPSP + Mg<br /> | **[[3w40]], [[3w41]] – BsPSP + Mg<br /> | ||
**[[3w42]], [[3w43]], [[3w44]] – BsPSP + Mn<br /> | **[[3w42]], [[3w43]], [[3w44]] – BsPSP + Mn<br /> | ||
| Line 35: | Line 36: | ||
**[[4ap9]] – ToPSP + pyridine derivative <br /> | **[[4ap9]] – ToPSP + pyridine derivative <br /> | ||
**[[4ij6]] – HtPSP (mutant) + phosphoserine<br /> | **[[4ij6]] – HtPSP (mutant) + phosphoserine<br /> | ||
| + | **[[5zr2]] – EhPSP (mutant) + phosphoserine <br /> | ||
*Phosphoserine phosphatase ternary complex | *Phosphoserine phosphatase ternary complex | ||
| + | **[[6q6j]] – hPSP + Ca + substrate analog<br /> | ||
| + | **[[6hyy]] – hPSP + PO4 + serine + <br /> | ||
**[[1l8o]] – hPSP (mutant) + serine + PO4<br /> | **[[1l8o]] – hPSP (mutant) + serine + PO4<br /> | ||
**[[1f5s]], [[1j97]], [[1l7m]] – MjPSP + PO4 + Mg<br /> | **[[1f5s]], [[1j97]], [[1l7m]] – MjPSP + PO4 + Mg<br /> | ||
| Line 43: | Line 47: | ||
**[[1l7p]] – MjPSP (mutant) + phosphoserine + PO4<br /> | **[[1l7p]] – MjPSP (mutant) + phosphoserine + PO4<br /> | ||
**[[5jlp]], [[5jlr]], [[5jma]] – MaPSP (mutant) + Mg + serine<br /> | **[[5jlp]], [[5jlr]], [[5jma]] – MaPSP (mutant) + Mg + serine<br /> | ||
| + | **[[8a1z]] – MaPSP + Mg + urea derivative<br /> | ||
| + | **[[8a21]] – MaPSP + Mg + phenylimidazole<br /> | ||
| + | **[[6m1x]] – EhPSP + PO4 + 3-phosphoglycerate<br /> | ||
| + | **[[7qpl]] – PSP + PO4 + Mg – ''Brucella melitensis''<br /> | ||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
3D structures of phosphoserine phosphatase
Updated on 29-August-2023
References
- ↑ NEUHAUS FC, BYRNE WL. Metabolism of phosphoserine. II. Purification and properties of O-phosphoserine phosphatase. J Biol Chem. 1959 Jan;234(1):113-21. PMID:13610904
- ↑ Jaeken J, Detheux M, Fryns JP, Collet JF, Alliet P, Van Schaftingen E. Phosphoserine phosphatase deficiency in a patient with Williams syndrome. J Med Genet. 1997 Jul;34(7):594-6. PMID:9222972
- ↑ Kim HY, Heo YS, Kim JH, Park MH, Moon J, Kim E, Kwon D, Yoon J, Shin D, Jeong EJ, Park SY, Lee TG, Jeon YH, Ro S, Cho JM, Hwang KY. Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase. J Biol Chem. 2002 Nov 29;277(48):46651-8. Epub 2002 Sep 3. PMID:12213811 doi:10.1074/jbc.M204866200