Bam complex
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
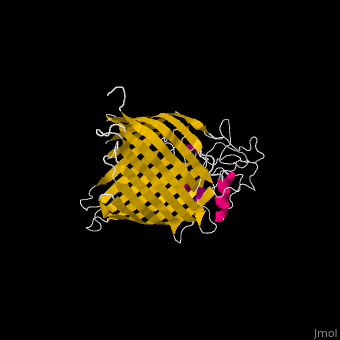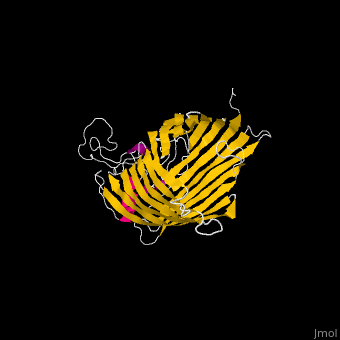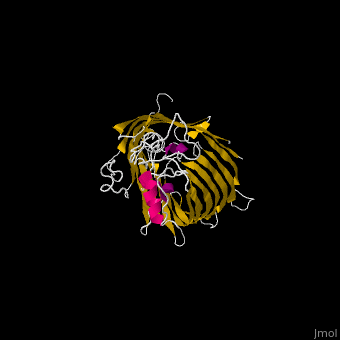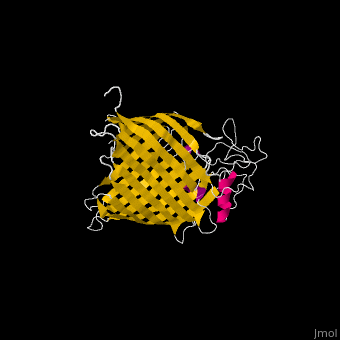
| - | <StructureSection load=' | + | <StructureSection load='' size='340' side='right' caption='E. coli BamA barrel domain (PDB code [[4n75]])' scene='70/706705/Cv/2'> |
== Function == | == Function == | ||
| Line 10: | Line 10: | ||
*'''BamA''' contains five structurally homologous POTRA (POlypeptide TRAnslocation associated) domains. The POTRA domain has a β-α-α-β-β conformation. BamA barrel and at least a subset of its POTRAs are essential for viability. BamA was found also in mitochondria and chloroplasts.<br /> | *'''BamA''' contains five structurally homologous POTRA (POlypeptide TRAnslocation associated) domains. The POTRA domain has a β-α-α-β-β conformation. BamA barrel and at least a subset of its POTRAs are essential for viability. BamA was found also in mitochondria and chloroplasts.<br /> | ||
*'''BamD''' is composed of multiple tetratricopeptide (TPR) repeats packed into a superhelical structure. TPR is a motif containing 2 antiparallel α-helices. TPRs are found in scaffold multiprotein complexes and are involved in protein-protein interactions. | *'''BamD''' is composed of multiple tetratricopeptide (TPR) repeats packed into a superhelical structure. TPR is a motif containing 2 antiparallel α-helices. TPRs are found in scaffold multiprotein complexes and are involved in protein-protein interactions. | ||
| - | |||
| - | </StructureSection> | ||
== 3D Structures of Bam complex == | == 3D Structures of Bam complex == | ||
| + | [[Bam complex 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009 Mar;7(3):206-14. doi: 10.1038/nrmicro2069. Epub 2009 Feb , 2. PMID:19182809 doi:http://dx.doi.org/10.1038/nrmicro2069