Peptidase T
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
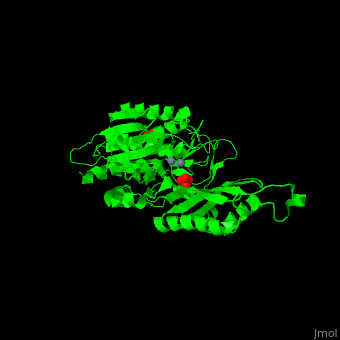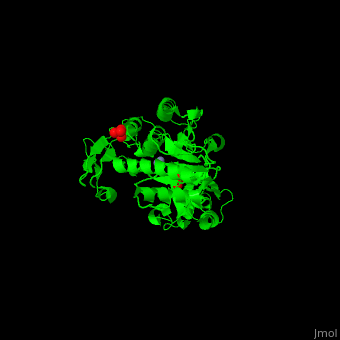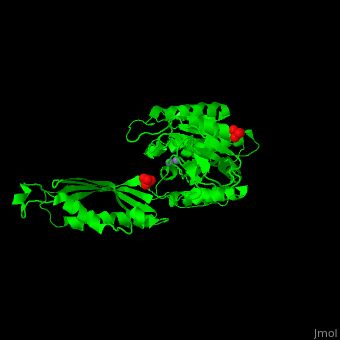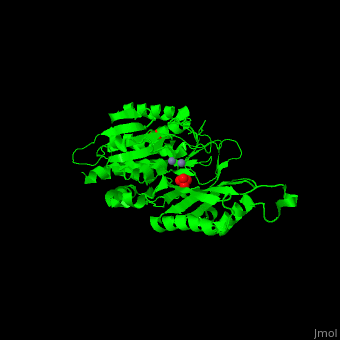
| - | <StructureSection load='1fno' size=' | + | <StructureSection load='1fno' size='350' side='right' scene='50/501403/Cv/1' caption='Monomer of the dimeric Salmonella typhimurium peptidase T complex with sulfate and Zn+2 ions (grey), [[1fno]]' pspeed='8'> |
'''Peptidase T''' (PT) removes the N-terminal peptide of tripeptides. PT is produced by bacteria. The tripeptides contain an N-terminal methionine, leucine or phenylalanine<ref>PMID:11856302</ref>. | '''Peptidase T''' (PT) removes the N-terminal peptide of tripeptides. PT is produced by bacteria. The tripeptides contain an N-terminal methionine, leucine or phenylalanine<ref>PMID:11856302</ref>. | ||
| - | *<scene name='50/501403/Cv/ | + | *<scene name='50/501403/Cv/3'>Zn coordinations sites</scene>. Water molecule are shown as red sphere. |
</StructureSection> | </StructureSection> | ||
==3D structures of peptidase T== | ==3D structures of peptidase T== | ||
Current revision
| |||||||||||
3D structures of peptidase T
Updated on 30-July-2019
1fno – PT – Salmonella typhymurium
3gb0 – PT – Bacillus cereus
3ife - PT – Bacillus anthracis
1vix – PT – Escherichia coli