1ie8
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
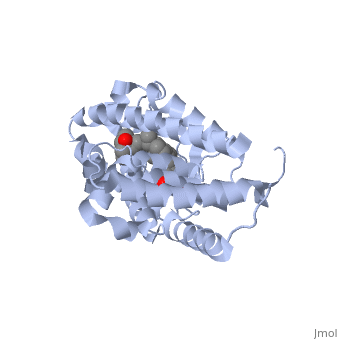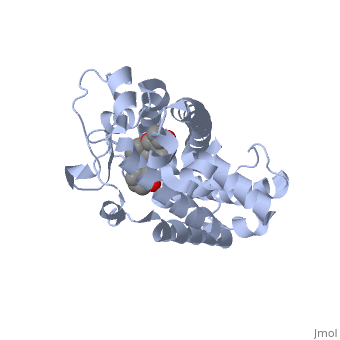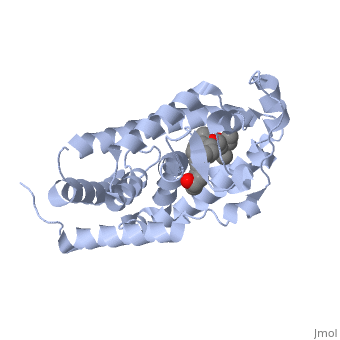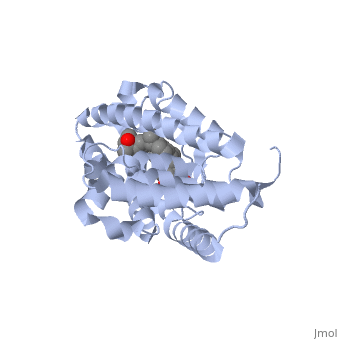
==Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060== | ==Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060== | ||
| - | <StructureSection load='1ie8' size='340' side='right' caption='[[1ie8]], [[Resolution|resolution]] 1.52Å' scene=''> | + | <StructureSection load='1ie8' size='340' side='right'caption='[[1ie8]], [[Resolution|resolution]] 1.52Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1ie8]] is a 1 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1ie8]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1IE8 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1IE8 FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=KH1:5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL'>KH1</scene> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.52Å</td></tr> |
| - | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=KH1:5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL'>KH1</scene></td></tr> | |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ie8 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ie8 OCA], [https://pdbe.org/1ie8 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ie8 RCSB], [https://www.ebi.ac.uk/pdbsum/1ie8 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ie8 ProSAT]</span></td></tr> |
</table> | </table> | ||
== Disease == | == Disease == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/VDR_HUMAN VDR_HUMAN] Defects in VDR are the cause of rickets vitamin D-dependent type 2A (VDDR2A) [MIM:[https://omim.org/entry/277440 277440]. A disorder of vitamin D metabolism resulting in severe rickets, hypocalcemia and secondary hyperparathyroidism. Most patients have total alopecia in addition to rickets.<ref>PMID:2849209</ref> <ref>PMID:8381803</ref> <ref>PMID:1652893</ref> <ref>PMID:2177843</ref> <ref>PMID:8106618</ref> <ref>PMID:8392085</ref> <ref>PMID:7828346</ref> <ref>PMID:8675579</ref> <ref>PMID:8961271</ref> <ref>PMID:9005998</ref> |
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/VDR_HUMAN VDR_HUMAN] Nuclear hormone receptor. Transcription factor that mediates the action of vitamin D3 by controlling the expression of hormone sensitive genes. Regulates transcription of hormone sensitive genes via its association with the WINAC complex, a chromatin-remodeling complex. Recruited to promoters via its interaction with the WINAC complex subunit BAZ1B/WSTF, which mediates the interaction with acetylated histones, an essential step for VDR-promoter association. Plays a central role in calcium homeostasis.<ref>PMID:16252006</ref> <ref>PMID:10678179</ref> <ref>PMID:15728261</ref> <ref>PMID:16913708</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
| - | <scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ie/1ie8_consurf.spt"</scriptWhenChecked> | + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ie/1ie8_consurf.spt"</scriptWhenChecked> |
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
| Line 22: | Line 22: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ie8 ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ie8 ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The crystal structures of the ligand-binding domain (LBD) of the vitamin D receptor complexed to 1alpha,25(OH)(2)D(3) and the 20-epi analogs, MC1288 and KH1060, show that the protein conformation is identical, conferring a general character to the observation first made for retinoic acid receptor (RAR) that, for a given LBD, the agonist conformation is unique, the ligands adapting to the binding pocket. In all complexes, the A- to D-ring moieties of the ligands adopt the same conformation and form identical contacts with the protein. Differences are observed only for the 17beta-aliphatic chains that adapt their conformation to anchor the 25-hydroxyl group to His-305 and His-397. The inverted geometry of the C20 methyl group induces different paths of the aliphatic chains. The ligands exhibit a low-energy conformation for MC1288 and a more strained conformation for the two others. KH1060 compensates this energy cost by additional contacts. Based on the present data, the explanation of the superagonist effect is to be found in higher stability and longer half-life of the active complex, thereby excluding different conformations of the ligand binding domain. | ||
| - | |||
| - | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.,Tocchini-Valentini G, Rochel N, Wurtz JM, Mitschler A, Moras D Proc Natl Acad Sci U S A. 2001 May 8;98(10):5491-6. PMID:11344298<ref>PMID:11344298</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1ie8" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
| - | *[[ | + | *[[Sandbox vdr|Sandbox vdr]] |
| + | *[[Vitamin D receptor 3D structures|Vitamin D receptor 3D structures]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Homo sapiens]] |
| - | [[Category: Mitschler | + | [[Category: Large Structures]] |
| - | [[Category: Moras | + | [[Category: Mitschler A]] |
| - | [[Category: Rochel | + | [[Category: Moras D]] |
| - | [[Category: Tocchini-Valentini | + | [[Category: Rochel N]] |
| - | [[Category: Wurtz | + | [[Category: Tocchini-Valentini G]] |
| - | + | [[Category: Wurtz JM]] | |
| - | + | ||
| - | + | ||
Current revision
Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060
| |||||||||||