Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | =='''Adhesin Competence Regulator'''== | + | =='''Adhesin Competence Regulator (AdcR)'''== |
<StructureSection load='3TGN' size='350' side='right' caption='[http://www.rcsb.org/pdb/explore/explore.do?structureId=3TGN Adhesin Competence Regulator (3TGN)]' scene=''> | <StructureSection load='3TGN' size='350' side='right' caption='[http://www.rcsb.org/pdb/explore/explore.do?structureId=3TGN Adhesin Competence Regulator (3TGN)]' scene=''> | ||
| Line 5: | Line 5: | ||
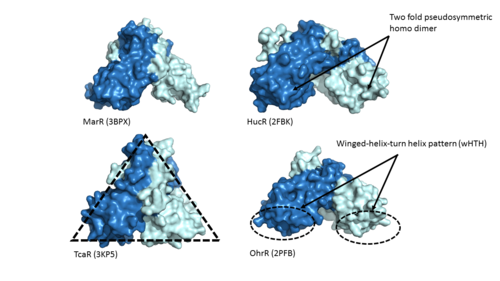
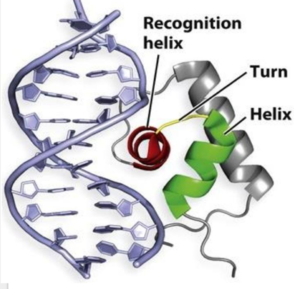
Adhesin Competence Regulator (<scene name='69/694230/Adcr_space_fill/1'>AdcR</scene>) is a transcriptional regulator that controls the activation of over seventy genes within the bacterium [https://en.wikipedia.org/wiki/Streptococcus_pneumoniae''Streptococcus pneumoniae''] <ref name="Sanson">DOI:10.1093/nar/gku1304 </ref> and is a member of the multiple antibiotic resistance regulator (MarR) protein family <ref> PMID: 23428319</ref>. Members of the Mar R protein family conserve a number of features including a general triangular shape, a two fold pseudosymmetric homodimer, and a winged helix-turn-helix pattern [https://en.wikipedia.org/wiki/Helix-turn-helix (wHTH)] which can be seen in Figure 1. AdcR exhibits these conserved features as well, while also exhibiting its own distinct features. | Adhesin Competence Regulator (<scene name='69/694230/Adcr_space_fill/1'>AdcR</scene>) is a transcriptional regulator that controls the activation of over seventy genes within the bacterium [https://en.wikipedia.org/wiki/Streptococcus_pneumoniae''Streptococcus pneumoniae''] <ref name="Sanson">DOI:10.1093/nar/gku1304 </ref> and is a member of the multiple antibiotic resistance regulator (MarR) protein family <ref> PMID: 23428319</ref>. Members of the Mar R protein family conserve a number of features including a general triangular shape, a two fold pseudosymmetric homodimer, and a winged helix-turn-helix pattern [https://en.wikipedia.org/wiki/Helix-turn-helix (wHTH)] which can be seen in Figure 1. AdcR exhibits these conserved features as well, while also exhibiting its own distinct features. | ||
| - | [[Image:MarR_protein_family_slide.png|500px|left|thumb|'''Figure 1'''. Proteins MarR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3bpx (3BPX)], HucR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2FBK (2FBK)], TcaR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3KP5 (3KP5)], and OhrR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2pfb (2PFB)] are pictured above with conserved features of the MarR protein family highlighted]] | + | [[Image:MarR_protein_family_slide.png|500px|left|thumb|'''Figure 1: MarR protein family features'''. Proteins MarR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3bpx (3BPX)], HucR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2FBK (2FBK)], TcaR [http://www.rcsb.org/pdb/explore/explore.do?structureId=3KP5 (3KP5)], and OhrR [http://www.rcsb.org/pdb/explore/explore.do?structureId=2pfb (2PFB)] are pictured above with conserved features of the MarR protein family highlighted]] |
In contrast with other members of the MarR family, AdcR is metal dependent. Zinc plays a vital role in organism homeostasis, acting as a [https://en.wikipedia.org/wiki/Cofactor_(biochemistry) co-factor] and a regulator of enzymatic activity. However zinc can lead to cell toxicity and deficiency of other vital metals that are also necessary for protein function <ref> DOI: 10.1021/cr900077w</ref>. Binding of Zinc allows AdcR to bind DNA and activate the transcription of high-affinity Zinc specific uptake transporters. The importance of AdcR in ''Streptococcus pneumoniae'' can be understood provided its ability to regulate zinc transfer proteins within the bacteria. | In contrast with other members of the MarR family, AdcR is metal dependent. Zinc plays a vital role in organism homeostasis, acting as a [https://en.wikipedia.org/wiki/Cofactor_(biochemistry) co-factor] and a regulator of enzymatic activity. However zinc can lead to cell toxicity and deficiency of other vital metals that are also necessary for protein function <ref> DOI: 10.1021/cr900077w</ref>. Binding of Zinc allows AdcR to bind DNA and activate the transcription of high-affinity Zinc specific uptake transporters. The importance of AdcR in ''Streptococcus pneumoniae'' can be understood provided its ability to regulate zinc transfer proteins within the bacteria. | ||
| Line 24: | Line 24: | ||
=== Binding Site 2 === | === Binding Site 2 === | ||
| - | <scene name='69/694230/Binding_site_2/4'>Binding site 2</scene> consists of a highly distorted tetrahedral geometry around the zinc ion. There are three amino acids involved in the binding of the zinc ion (C30, E41, and E107) as well as a water molecule (shown as a red sphere). | + | <scene name='69/694230/Binding_site_2/4'>Binding site 2</scene> consists of a highly distorted tetrahedral geometry around the zinc ion. There are three amino acids involved in the binding of the zinc ion (C30, E41, and E107) as well as a water molecule (shown as a red sphere). When Cys30 in binding site 2 is mutated to an alanine, it has no effect on the ability of the protein to bind DNA <ref name="guerra" />. Therefore, binding site 2 has no significant role in the ability of AdcR to bind to DNA and AdcR is still able to function with no zinc bound present in binding site 2. In fact, the presence of binding site 2 may simply be due to an excess of zinc during the crystallization process. |
=== Hydrogen Bond Network === | === Hydrogen Bond Network === | ||
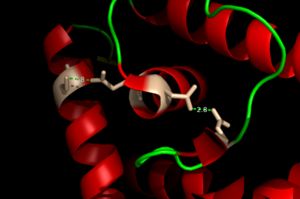
| - | The binding of zinc metals creates a hydrogen bond network within the protein that connects the metal binding sites and the [https://en.wikipedia.org/wiki/DNA-binding_domain DNA binding domain]. The <scene name='69/694230/Hydrogen_bonding_1/5'>hydrogen bond network</scene> (<scene name='69/694230/Hydrogen_bonding_2/5'>with measurements</scene>) is represented by each atom type in the 3D model. The hydrogen bond network is characteristic of the MarR family as a whole. More importantly, the hydrogen bonding network connects the metal binding pockets to the α4 helix also known as the recognition helix. <scene name='69/694230/Recognition_helix/3'>Several residues</scene> in the recognition helix recognize a sequence of DNA that is unknown at the moment; however, scientists are aware that the hydrogen bond network acts as an allosteric activator for the protein to bind DNA. The hydrogen bond network connects the α2 and α4 helix via hydrogen bonding between specific residues. After zinc is bound, a glutamate (E24) residue from a random coil accepts a hydrogen bond from the carboxamide end of an asparagine (N38) residue from the α2 helix. Then, a glutamine (Q40) residue from α2 helix accepts a hydrogen bond from a serine (S74) residue from the α4 helix <ref name="guerra" />. The binding of zinc allows for these conformational changes that induces the binding of DNA in order to activate genes. | ||
[[Image:H Bonding of DNA.png|300 px|left|thumb|'''Figure 4'''. The Hydrogen Bonding Network is shown with dotted green lines approximately 2.8 angstroms between residues.]] | [[Image:H Bonding of DNA.png|300 px|left|thumb|'''Figure 4'''. The Hydrogen Bonding Network is shown with dotted green lines approximately 2.8 angstroms between residues.]] | ||
| + | The binding of zinc metals creates a hydrogen bond network (Figure 4) within the protein that connects the metal binding sites and the [https://en.wikipedia.org/wiki/DNA-binding_domain DNA binding domain]. The <scene name='69/694230/Hydrogen_bonding_1/5'>hydrogen bond network</scene> (<scene name='69/694230/Hydrogen_bonding_2/5'>with measurements</scene>) (residues in stick structures, colored by atom type) is characteristic of the MarR family as a whole and connects the metal binding pockets to the α4 helix also known as the DNA recognition helix. <scene name='69/694230/Recognition_helix/3'>Several residues</scene> in this helix recognize the DNA ligand. The hydrogen bond network connects the α2 and α4 helices via hydrogen bonding between specific residues. After zinc is bound, a glutamate (E24) residue from a random coil accepts a hydrogen bond from the carboxamide end of an asparagine (N38) residue from the α2 helix. A glutamine (Q40) residue from α2 helix accepts a hydrogen bond from a serine (S74) residue from the α4 helix <ref name="guerra" />. | ||
| - | == ''' | + | == '''Clinical Relevance''' == |
| - | Streptococcus pneumoniae | + | ''Streptococcus pneumoniae'' is a significant pathogenic bacterium. Although asymptomatic in healthy individuals, ''S. pneumoniae'' can lead to Bronchitis, meningitis conjunctivitis, or brain abscesses in those with weaker immune systems. Host regulation of zinc is often used to combat pathogens such as ''S. pneumoniae'' <ref name="Sanson" />. A better understanding of AdcR, the regulator that controls the transcription of zinc specific uptake transporters, could help to illuminate better mechanism for combating not only ''S. pneumoniae'', but other comparable bacteria. |
</StructureSection> | </StructureSection> | ||
Current revision
Adhesin Competence Regulator (AdcR)
| |||||||||||
References
- ↑ 1.0 1.1 Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 2015 Jan;43(1):418-32. doi: 10.1093/nar/gku1304. Epub 2014 Dec, 15. PMID:25510500 doi:http://dx.doi.org/10.1093/nar/gku1304
- ↑ Grove A. MarR family transcription factors. Curr Biol. 2013 Feb 18;23(4):R142-3. doi: 10.1016/j.cub.2013.01.013. PMID:23428319 doi:http://dx.doi.org/10.1016/j.cub.2013.01.013
- ↑ Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009 Oct;109(10):4644-81. doi: 10.1021/cr900077w. PMID:19788177 doi:http://dx.doi.org/10.1021/cr900077w
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Guerra AJ, Dann CE, Giedroc DP. Crystal Structure of the Zinc-Dependent MarR Family Transcriptional Regulator AdcR in the Zn(II)-Bound State. J Am Chem Soc. 2011 Nov 21. PMID:22085181 doi:10.1021/ja2080532
- ↑ 5.0 5.1 Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UM, Scott RA, Winkler ME, Giedroc DP. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol. 2010 Oct 22;403(2):197-216. doi: 10.1016/j.jmb.2010.08.030. Epub 2010, Sep 8. PMID:20804771 doi:http://dx.doi.org/10.1016/j.jmb.2010.08.030