User:Rafael Romero Becerra/Sandbox 1
From Proteopedia
< User:Rafael Romero Becerra(Difference between revisions)
| (27 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='2pmw' size='350' side='right' caption=' | + | <StructureSection load='2pmw' size='350' side='right' caption='PCSK9' scene='77/774675/Pcsk9-domains/14'> |
| - | '''Pro-protein convertase subtilisin/kexin type 9 (PCSK9)''' the ninth known member of the mammalian serine proprotein convertase (PC) family, and plays an important role in low density lipoproteins (LDL) metabolism. Once secreted, PCSK9 binds LDL receptors (LDLRs), targeting them toward intracellular degradation through an endosomal/lysosomal route. Inhibition of PCSK9 can reduce LDLRs degradation and increase the expression of LDLRs in the cell surface, resulting in an enhanced recycling of LDLRs and a reduction in the levels of LDL cholesterol. Hence, inhibitors of PCSK9 suppose a promising therapeutic strategy for the treatment of hypercholesterolemia. | + | '''Pro-protein convertase subtilisin/kexin type 9 (PCSK9)''' is the ninth known member of the mammalian serine proprotein convertase (PC) family, and plays an important role in low density lipoproteins (LDL) metabolism. Once secreted, PCSK9 binds LDL receptors (LDLRs), targeting them toward intracellular degradation through an endosomal/lysosomal route. Inhibition of PCSK9 can reduce LDLRs degradation and increase the expression of LDLRs in the cell surface, resulting in an enhanced recycling of LDLRs and a reduction in the levels of LDL cholesterol. Hence, inhibitors of PCSK9 suppose a promising therapeutic strategy for the treatment of hypercholesterolemia. |
== Discovery of PCSK9 == | == Discovery of PCSK9 == | ||
| - | PCSK9 was first described as neural apoptosis-regulated convertase 1 (NARC-1) in studies of cerebral neuron apoptosis, suggesting that it could be implicated in the differentiation of cortical neurons <ref>DOI 10.1073/pnas.0335507100</ref>. Concomitant and following studies in patients with familiar hypercholesterolemia revealed the clinical importance of PCSK9, showing that patients with gain-of-function mutations presented increased levels of cholesterol in plasma (i.e. hypercholesterolemia) due to reduced expression of LDLRs. In contrast, loss-of-function variants of PSCK9 are associated with a reduction of LDL cholesterol levels and a lower risk of cardiovascular disease. The role of PSCK9 in LDLRs and cholesterol metabolism has been confirmed in animal models. Thus, mice overexpressing PCSK9 show a reduction in the expression of hepatic LDLRs and hypercholesterolemia, whereas knockout mice for PCSK9 present decreased levels of plasmatic LDL cholesterol because of increased expression of LDLRs <ref name=Abifadel>DOI 10.1002/humu.20882</ref><ref name=Hess>DOI 10.1146/annurev-med-042716-091351</ref>. | + | PCSK9 was first described as neural apoptosis-regulated convertase 1 (NARC-1) in studies of cerebral neuron apoptosis, suggesting that it could be implicated in the differentiation of cortical neurons <ref name=Seidah>DOI 10.1073/pnas.0335507100</ref>. Concomitant and following studies in patients with familiar hypercholesterolemia revealed the clinical importance of PCSK9, showing that patients with gain-of-function mutations presented increased levels of cholesterol in plasma (i.e. hypercholesterolemia) due to reduced expression of LDLRs. In contrast, loss-of-function variants of PSCK9 are associated with a reduction of LDL cholesterol levels and a lower risk of cardiovascular disease. The role of PSCK9 in LDLRs and cholesterol metabolism has been confirmed in animal models. Thus, mice overexpressing PCSK9 show a reduction in the expression of hepatic LDLRs and hypercholesterolemia, whereas knockout mice for PCSK9 present decreased levels of plasmatic LDL cholesterol because of increased expression of LDLRs <ref name=Abifadel>DOI 10.1002/humu.20882</ref><ref name=Hess>DOI 10.1146/annurev-med-042716-091351</ref>. |
== Gene expression and synthesis of PCSK9 == | == Gene expression and synthesis of PCSK9 == | ||
| + | |||
| + | PCSK9 is the ninth known member of the mammalian subtilisin (S8) serine proprotein convertase (PC) family that carries out the proteolytic maturation of secretory proteins such as neuropeptides, prohormones and cytokines. Humans have nine different PCs that can be divided between S8A and S8B subfamilies. PCSK9 is classified in subfamily S8A <ref name=Piper>DOI 10.1016/j.str.2007.04.004 </ref>. | ||
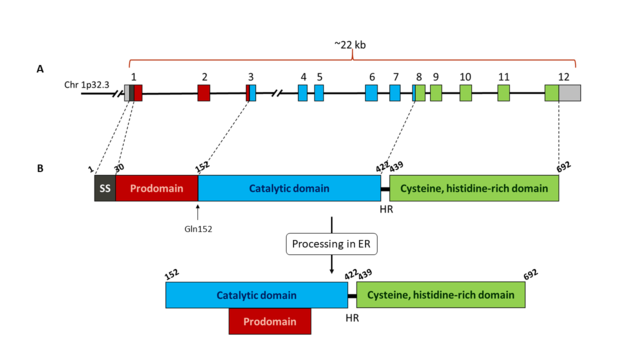
| - | + | The human gene for PCSK9 is 22 kb length and it is located in chromosome 1p32.3. It contains 11 introns and 12 exons that encode the 692 amino acids of the enzyme. The sequence of the protein is characterized by a signal sequence (amino acids 1-30), a prodomain (amino acids 31-152), and a catalytic domain, followed by a C-terminal region of 243 amino acids which is rich in cysteine and histidine residues. PCSK9 is mainly expressed in the liver, intestine and kidney, and it can also be in the nervous system. It is synthesized as a precursor of ~74 kDa that is processed in the endoplasmic reticulum (ER) where it undergoes cleavage of its signal peptide and intramolecular autocatalytic cleavage producing a ~60-kDa catalytic fragment. The autocatalysis of the zymogen takes place between Gln152 and Ser153 <ref name=Naureckiene>PMID:14622975</ref>. This cleavage is necessary for transport from ER to the Golgi body and for secretion. The cleaved prodomain of ~14 kDa remains associated with the catalytic domain, which is unique to PCSK9. This facilitates protein folding, permits the mature protein to move from ER into the secretory pathway and regulates the catalytic activity of the enzyme by blocking the access to the catalytic site <ref name=Abifadel /><ref name=Hess />.[[Image:PCSK9_domains2.png|thumb|center|640x360px| Schematic representation of PCSK9 gene (A) and protein (B). A: exons of PCSK9 gene are shown as coloured boxes. Each colour corresponds to a different domain of the protein. B: Representation of PCSK9 protein domains: signal sequence (SS, black), prodomain (red), catalytic domain (blue) and Cys, His rich C-terminal domain (green). The autocatalytic cleavage site at Gln152 is indicated with an arrow. Once the protein has been processed in the endoplasmic reticulum (ER), the prodomain remains bound to the catalytic domain. The catalytic domain is linked to the C-terminal domain through a 18 amino acids hinge region (HR). Numbers above each domain indicate the amino acid number of the protein sequence.]] | |
| - | + | ||
| - | + | PCSK9 can be found in plasma in two forms: the mature and secreted form of ~60 kDa, and as an inactivated fragment of ~53 kDa produced by the cleavage of the mature form at the motive RFHR218↓ by other proprotein convertases, mainly furin and/or PC5/6A <ref name=Benjannet>DOI 10.1074/jbc.M606495200</ref>. | |
| - | + | In humans, PCSK9 circulates in plasma in a phosphorylated state and it has been shown that it is phosphorylated at the Ser47 and Ser688 by a Golgi casein kinase-like kinase ex vivo. This phosphorylation might be important to protect the propeptide against proteolysis <ref name=Dewpura>DOI 10.1111/j.1742-4658.2008.06495.x</ref>. | |
| - | PCSK9 can be found in plasma in two forms: the mature and secreted form of ~60 kDa, and as an inactivated fragment of ~53 kDa produced by the cleavage of the mature form at the motive RFHR218↓ by other proprotein convertases, mainly furin and/or PC5/6A <ref>DOI 10.1074/jbc.M606495200</ref>. | + | |
| - | In humans, PCSK9 circulates in plasma in a phosphorylated state and it has been shown that it is phosphorylated at the Ser47 and Ser688 by a Golgi casein kinase-like kinase ex vivo. This phosphorylation might be important to protect the propeptide against proteolysis <ref>DOI 10.1111/j.1742-4658.2008.06495.x</ref>. | + | |
=== Regulation of ''PCSK9'' gene expression === | === Regulation of ''PCSK9'' gene expression === | ||
| - | The expression of PCSK9 and LDLR genes are regulated by a common pathway. Thus, when there is reduction in the cholesterol cell content, the transcription of both genes is induced through the sterol regulatory element-binding protein (SREBP) <ref name=Costet> DOI 10.1074/jbc.M508582200</ref>. Additionally, there are evidences that statins also upregulate the expression of PCSK9 gene <ref>DOI 10.1161/01.ATV.0000134621.14315.43</ref>. | + | The expression of PCSK9 and LDLR genes are regulated by a common pathway. Thus, when there is reduction in the cholesterol cell content, the transcription of both genes is induced through the sterol regulatory element-binding protein (SREBP) <ref name=Costet> DOI 10.1074/jbc.M508582200</ref>. Additionally, there are evidences that statins also upregulate the expression of PCSK9 gene <ref name=Dubuc>DOI 10.1161/01.ATV.0000134621.14315.43</ref>. |
Another factor that affects the expression of PCSK9 is the nutritional status. It has been shown that fasting decreases the expression of hepatic PCSK9 in mice, and that the level of PCSK9 gene expression is recovered upon refeeding. Furthermore, insulin also upregulates the hepatic PCSK9 gene expression, suggesting a possible role of PCSK9 in insulin-related diseases like type 2 diabetes. The liver X receptor (LXR) and sterol regulatory element-binding protein 1c (SREBP-1c) would be involved in the pathway that regulates the expression of PCSK9 according to the nutritional status <ref name=Costet />. | Another factor that affects the expression of PCSK9 is the nutritional status. It has been shown that fasting decreases the expression of hepatic PCSK9 in mice, and that the level of PCSK9 gene expression is recovered upon refeeding. Furthermore, insulin also upregulates the hepatic PCSK9 gene expression, suggesting a possible role of PCSK9 in insulin-related diseases like type 2 diabetes. The liver X receptor (LXR) and sterol regulatory element-binding protein 1c (SREBP-1c) would be involved in the pathway that regulates the expression of PCSK9 according to the nutritional status <ref name=Costet />. | ||
| - | == | + | == Function == |
| - | < | + | |
| + | Although the first role suggested for PCSK9 was neuronal differentiation <ref name=Seidah />, later it was found that PCSK9 is involved in LDL cholesterol metabolism. | ||
| + | The best-characterized role of the mature and secreted form of PCSK9 (the ~60 kDa cleaved enzyme with the ~14-kDa prodomain associated to the catalytic domain) is targeting LDLRs for degradation in the liver. The catalytic subunit binds the epidermal growth factor-A (EGF-A) domain of the LDLR at the hepatocyte cell surface leading to LDLR internalization and degradation. | ||
| + | Once LDL cholesterol binds LDLR, it enters the cell through clathrin-coated vesicles. After internalization, the acidic pH of endosomes disrupts the association of LDL cholesterol from its receptor. LDL particles remain within the endosome while a recycling vesicle returns the LDLR to the cell surface. Endosomes containing LDL cholesterol fuse with lysomes where LDL is degraded and cholesterol esters are hydrolyzed. The free cholesterol is then distributed to other cellular compartments. At the hepatocyte cell surface, the catalytic domain of PCSK9 can also bind LDLR. The complex is the internalized via clathrin-coated vesicles. Within the endosome, the affinity of PCSK9 for the LDLR is enhanced due to the low pH, preventing the recycling of the receptor to the cell surface. The complex is then directed to the lysosome, where both components, LDLR and PCSK9, are degraded <ref name=Burke>DOI 10.1146/annurev-pharmtox-010716-104944</ref><ref name=Hess />. In addition, in vitro studies in hepatocytes suggest that PCSK9 might also enhance intracellular LDLR degradation prior to its secretion. When PCSK9 binds to LDLR within the Golgi complex, there is an increase in the traffic of LDLR bound to PCSK9 from the trans Golgi network to lysosomes for degradation, instead of directing the receptors to the cell surface <ref name=Poirier>DOI 10.1074/jbc.M109.037085</ref>. It has been suggested that PCSK9 might also induced LDLR degradation by ubiquitination of the receptor <ref name=Chen>DOI 10.1016/j.bbrc.2011.10.110</ref>. | ||
| - | < | + | In addition to binding to LDLRs in the liver, PCSK9 has other less characterized roles in different tissues. In the small intestine, PCSK9 might regulate the production of triglyceride-rich apolipoprotein B and might also regulate transintestinal fecal cholesterol excretion. Furthermore, PCSK9 has been suggested to regulate the expression of very-low-density lipoprotein receptors (VLDLRs) in adipose tissue and the ApoE receptor 2 receptor in the brain, by means of a similar mechanism to the LDLR. PCSK9 binds VLDLR and ApoE receptor 2, finally resulting in the lysosomal degradation of the receptors. By modulation of ApoE receptor 2 and related anti-apoptotic signaling pathways, PSCK9 might regulate neuronal apoptosis. Endocrine pancreatic cells also expressed PCSK9, but it is not known the role of PCSK9 in pancreatic cells <ref name=Hess />. PCSK9 also inhibits epithelial Na<sup>+</sup> channel (ENaC)-mediated Na<sup>+</sup> absorption by reducing ENaC surface expression, mainly by an increased proteasomal degradation. By reducing ENaC channel number, PCSK9 might modulate epithelial Na<sup>+</sup> absorption, which is a major contributor to blood pressure control <ref name=Sharotri>DOI 10.1074/JBC.M112.363382</ref>. |
| - | + | == PCSK9 and hypercholesterolemia == | |
| - | + | High serum levels of LDL cholesterol are strongly associated with a higher risk of developing cardiovascular disease (CVD), which is the leading cause of death worldwide. | |
| + | People with familial hypercholesterolemia present mutations that cause defects in hepatic cholesterol clearance, leading to increased levels of LDL cholesterol in plasma and, therefore, increased risk of CVD. Previously, mutations in LDL receptor and apolipoprotein B (an essential component of LDL particles which interacts with LDLR) were linked to familial hypercholesterolemia. The studies performed during the last years have identified the gene encoding PCSK9 as a third locus related to familial hypercholesterolemia. There is a link between PCSK9 function and LDL cholesterol serum levels <ref name=Abifadel />. Thus, gain of function mutations in PCSK9 cause an infrequent form of familial hypercholesterolemia, while loss of function mutations are associated with hypocholesterolemia and reduced risk of CVD. | ||
| + | There is a relationship between lowering LDL cholesterol and reduction in risk of CVD. Hence, one of the main strategies to prevent CVD is reducing LDL cholesterol serum levels. The most used treatment to reduce LDL levels is statins, which inhibits 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase, a key enzyme in the synthesis of cholesterol. However, these drugs have some adverse effects including myopathy, rhabdomyolysis and hemorrhagic stroke, and some patients do not respond properly to the treatment. PCSK9 has become a promising target to treat hypercholesterolemia when statins are not a proper option. Inhibition of PCSK9 avoids LDLR degradation resulting in an increase of LDLR at the hepatocyte cell surface, enhancing the uptake of LDL cholesterol and lowering LDL cholesterol circulating levels. Several monoclonal antibodies against PCSK9 are being already used in phase III clinical trials, and there is an active research in other strategies to inhibit PCSK9 ([[#PCSK9 as a therapeutic target|discussed below]]). | ||
| - | + | == PCSK9/LDLR complex == | |
| - | + | The '''interaction''' between PCSK9 and LDL receptor can be explained by a two-step model. In the first step, the <scene name='77/774675/Pro_cat_domain/1'>Pro-Cat domain</scene> of PCSK9 initiates contact with EGF-A of the LDL receptor at neutral pH. An <scene name='77/774675/Beta_better/2'>antiparallel β-sheet</scene> is formed between residues 377– 379 of PCSK9 and residues 308–310 of EGF-A. In the second step, the complex is internalized and exposure to the low pH environment of the endosome, increasing '''positive charge density''' in the CT domain (owing to side chain ionization of its numerous surface-exposed <scene name='77/774675/His/1'>histidines</scene> residues (7–9)). This change in charge density promotes the interaction of the CT domain of PCSK9 with the Ligand-Binding domain of LDLR, which impairs the ability of the receptor to adopt a recycling-competent conformation and promote trafficking of the PCSK9-LDLR complex to the lysosome. Interestingly, it has been observed that there are '''two populations of binding sites''' at pH 5.4, a high affinity | |
| + | binding site (Kd~1 nM) and a lower affinity binding site (Kd~50 nM). Whether these may correspond to Pro-Cat domain binding to EGF-A and CT domain binding to the LBD, respectively, remains to be determined<ref>PMID:21149300</ref>. | ||
| + | In the first step, most of the contacts between PCSK9 and LDLR occur between the PCSK9 catalytic domain and the LDLR EGF-A domain. But the PCSK9 prodomain makes also van der Waals contacts with the LDLR beta propeller, creating a <scene name='77/774675/Second_binding_site/3'>second binding site</scene>. And it has been described that some mutations that affect these contacts are associated with familial hypercholesterolemia. Particularly, the PCSK9 S127R GOF mutation maps to this region, suggesting that extra contacts with the beta propeller might underlie this phenotype increasing the affinity<ref>PMID:22081141</ref>. | ||
| + | Furthermore, the structure showed that the PCSK9 CTD does not contact the LDLR and is solvent exposed. This feature is consistent with previous studies in which it is showed that CTD deletion does not affect PCSK9/LDLR binding at neutral pH. It is believed that CTD is binding to a cell surface co-receptor, but it is still<ref>PMID:22081141</ref>. | ||
== Kinetics of PCSK9 == | == Kinetics of PCSK9 == | ||
| - | Under normal conditions, PCSK9 has a half-life in plasma of approximately 5 minutes. It has been showed that in humans and mice, LDLR is a major regulator for PCSK9 levels and clearance, therefore in the presence of an additional copy of LDLR in the liver (induced by transgenic expression) reduces the half-life of PCSK9 by 50%, to 2.9 minutes, whereas in the absence of LDLR, the half-life of PCSK9 in serum is prolonged between 3–10 times above normal. | + | Under '''normal conditions''', PCSK9 has a half-life in plasma of approximately 5 minutes. It has been showed that in humans and mice, LDLR is a major regulator for PCSK9 levels and clearance, therefore in the presence of an '''additional copy of LDLR''' in the liver (induced by transgenic expression) reduces the half-life of PCSK9 by 50%, to 2.9 minutes, whereas in the absence of LDLR, the half-life of PCSK9 in serum is prolonged between 3–10 times above normal. |
| - | The kinetics of wild-type (WT) PCSK9 binding to LDLR shows Kd | + | The kinetics of wild-type (WT) PCSK9 binding to LDLR shows Kd values that range from 90 to 840 nM at '''neutral pH''', and its affinity to LDLR becomes ∼100-fold higher at '''lower pH''' with Kd values ranging from 1 to 8 nM. PCSK9 binding to LDLR has been described as '''biphasic''', with a first rapid phase characterized by a half-time of 6.6 minutes, which accounts for 35% of the equilibrium binding and a second slow phase whose half-time is 94 minutes. Similarly, 25% of the PCSK9 bound to LDLR dissociates during the rapid phase with a half-time of 19 minutes, while the remaining PCSK9 dissociates slowly with a half-time of 297 minutes. |
| - | + | Despite the rapid binding of PCSK9 and internalization of LDLR, PCSK9-mediated degradation of LDLR in vitro has only been observed after several hours. It was further shown that, at least in mice, PCSK9 remains intact in the liver for up to 4 hours after its internalization, thus suggesting that other events might be required in order to allow PCSK9-mediated degradation of LDLR (or LDLR mediated degradation of PCSK9)<ref>PMID:26345307</ref>. | |
| - | + | ||
| - | Despite the rapid binding of PCSK9 and internalization of LDLR | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| Line 143: | Line 137: | ||
The ''inhibitory activity'' is based in the introduction of modifications in the protein extension to reduce the favourable interactions of PCSK9 with the LDLR-EGFA. The mechanism is based the presence of certain aminoacids that enable the ability to extend toward the EGFA binding site. In this regard, antagonism is based in the steric clash of EGFA residues Leu 298, Asp299 and Asn 300 with the Pro 5 residue from the peptide. Furthermore, the presence of a common phenylalanine/tyrosine–proline–glycine (FPG/YPG) common domain in the extension peptide adopting a β-turn conformation also antagonize the binding of LDLR receptor<ref name=Zhang2017 />. | The ''inhibitory activity'' is based in the introduction of modifications in the protein extension to reduce the favourable interactions of PCSK9 with the LDLR-EGFA. The mechanism is based the presence of certain aminoacids that enable the ability to extend toward the EGFA binding site. In this regard, antagonism is based in the steric clash of EGFA residues Leu 298, Asp299 and Asn 300 with the Pro 5 residue from the peptide. Furthermore, the presence of a common phenylalanine/tyrosine–proline–glycine (FPG/YPG) common domain in the extension peptide adopting a β-turn conformation also antagonize the binding of LDLR receptor<ref name=Zhang2017 />. | ||
| - | == Disease == | ||
| - | |||
| - | == Relevance == | ||
| - | |||
| - | == Structural highlights == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):928-33. Epub 2003 Jan 27. PMID:12552133 doi:http://dx.doi.org/10.1073/pnas.0335507100
- ↑ 2.0 2.1 2.2 Abifadel M, Rabes JP, Devillers M, Munnich A, Erlich D, Junien C, Varret M, Boileau C. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease. Hum Mutat. 2009 Apr;30(4):520-9. doi: 10.1002/humu.20882. PMID:19191301 doi:http://dx.doi.org/10.1002/humu.20882
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Hess CN, Low Wang CC, Hiatt WR. PCSK9 Inhibitors: Mechanisms of Action, Metabolic Effects, and Clinical Outcomes. Annu Rev Med. 2017 Nov 2. doi: 10.1146/annurev-med-042716-091351. PMID:29095667 doi:http://dx.doi.org/10.1146/annurev-med-042716-091351
- ↑ Piper DE, Jackson S, Liu Q, Romanow WG, Shetterly S, Thibault ST, Shan B, Walker NP. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure. 2007 May;15(5):545-52. PMID:17502100 doi:http://dx.doi.org/10.1016/j.str.2007.04.004
- ↑ Naureckiene S, Ma L, Sreekumar K, Purandare U, Lo CF, Huang Y, Chiang LW, Grenier JM, Ozenberger BA, Jacobsen JS, Kennedy JD, DiStefano PS, Wood A, Bingham B. Functional characterization of Narc 1, a novel proteinase related to proteinase K. Arch Biochem Biophys. 2003 Dec 1;420(1):55-67. PMID:14622975
- ↑ Benjannet S, Rhainds D, Hamelin J, Nassoury N, Seidah NG. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J Biol Chem. 2006 Oct 13;281(41):30561-72. Epub 2006 Aug 15. PMID:16912035 doi:http://dx.doi.org/10.1074/jbc.M606495200
- ↑ Dewpura T, Raymond A, Hamelin J, Seidah NG, Mbikay M, Chretien M, Mayne J. PCSK9 is phosphorylated by a Golgi casein kinase-like kinase ex vivo and circulates as a phosphoprotein in humans. FEBS J. 2008 Jul;275(13):3480-93. doi: 10.1111/j.1742-4658.2008.06495.x. Epub, 2008 May 22. PMID:18498363 doi:http://dx.doi.org/10.1111/j.1742-4658.2008.06495.x
- ↑ 8.0 8.1 Costet P, Cariou B, Lambert G, Lalanne F, Lardeux B, Jarnoux AL, Grefhorst A, Staels B, Krempf M. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J Biol Chem. 2006 Mar 10;281(10):6211-8. doi: 10.1074/jbc.M508582200. Epub 2006, Jan 6. PMID:16407292 doi:http://dx.doi.org/10.1074/jbc.M508582200
- ↑ Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, Prat A. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004 Aug;24(8):1454-9. doi:, 10.1161/01.ATV.0000134621.14315.43. Epub 2004 Jun 3. PMID:15178557 doi:http://dx.doi.org/10.1161/01.ATV.0000134621.14315.43
- ↑ Burke AC, Dron JS, Hegele RA, Huff MW. PCSK9: Regulation and Target for Drug Development for Dyslipidemia. Annu Rev Pharmacol Toxicol. 2017 Jan 6;57:223-244. doi:, 10.1146/annurev-pharmtox-010716-104944. Epub 2016 Aug 8. PMID:27575716 doi:http://dx.doi.org/10.1146/annurev-pharmtox-010716-104944
- ↑ Poirier S, Mayer G, Poupon V, McPherson PS, Desjardins R, Ly K, Asselin MC, Day R, Duclos FJ, Witmer M, Parker R, Prat A, Seidah NG. Dissection of the endogenous cellular pathways of PCSK9-induced low density lipoprotein receptor degradation: evidence for an intracellular route. J Biol Chem. 2009 Oct 16;284(42):28856-64. doi: 10.1074/jbc.M109.037085. Epub, 2009 Jul 27. PMID:19635789 doi:http://dx.doi.org/10.1074/jbc.M109.037085
- ↑ Chen Y, Wang H, Yu L, Yu X, Qian YW, Cao G, Wang J. Role of ubiquitination in PCSK9-mediated low-density lipoprotein receptor degradation. Biochem Biophys Res Commun. 2011 Nov 25;415(3):515-8. doi:, 10.1016/j.bbrc.2011.10.110. Epub 2011 Nov 2. PMID:22074827 doi:10.1016/j.bbrc.2011.10.110
- ↑ Sharotri V, Collier DM, Olson DR, Zhou R, Snyder PM. Regulation of epithelial sodium channel trafficking by proprotein convertase subtilisin/kexin type 9 (PCSK9). J Biol Chem. 2012 Jun 1;287(23):19266-74. doi: 10.1074/jbc.M112.363382. Epub 2012, Apr 9. PMID:22493497 doi:10.1074/jbc.M112.363382
- ↑ Yamamoto T, Lu C, Ryan RO. A two-step binding model of PCSK9 interaction with the low density lipoprotein receptor. J Biol Chem. 2011 Feb 18;286(7):5464-70. doi: 10.1074/jbc.M110.199042. Epub 2010 , Dec 11. PMID:21149300 doi:http://dx.doi.org/10.1074/jbc.M110.199042
- ↑ Lo Surdo P, Bottomley MJ, Calzetta A, Settembre EC, Cirillo A, Pandit S, Ni YG, Hubbard B, Sitlani A, Carfi A. Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO Rep. 2011 Dec 1;12(12):1300-5. doi: 10.1038/embor.2011.205. PMID:22081141 doi:http://dx.doi.org/10.1038/embor.2011.205
- ↑ Lo Surdo P, Bottomley MJ, Calzetta A, Settembre EC, Cirillo A, Pandit S, Ni YG, Hubbard B, Sitlani A, Carfi A. Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO Rep. 2011 Dec 1;12(12):1300-5. doi: 10.1038/embor.2011.205. PMID:22081141 doi:http://dx.doi.org/10.1038/embor.2011.205
- ↑ Giunzioni I, Tavori H. New developments in atherosclerosis: clinical potential of PCSK9 inhibition. Vasc Health Risk Manag. 2015 Aug 24;11:493-501. doi: 10.2147/VHRM.S74692., eCollection 2015. PMID:26345307 doi:http://dx.doi.org/10.2147/VHRM.S74692
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 El Khoury P, Elbitar S, Ghaleb Y, Khalil YA, Varret M, Boileau C, Abifadel M. PCSK9 Mutations in Familial Hypercholesterolemia: from a Groundbreaking Discovery to Anti-PCSK9 Therapies. Curr Atheroscler Rep. 2017 Oct 17;19(12):49. doi: 10.1007/s11883-017-0684-8. PMID:29038906 doi:http://dx.doi.org/10.1007/s11883-017-0684-8
- ↑ 19.0 19.1 Zhang Y, Ultsch M, Skelton NJ, Burdick DJ, Beresini MH, Li W, Kong-Beltran M, Peterson A, Quinn J, Chiu C, Wu Y, Shia S, Moran P, Di Lello P, Eigenbrot C, Kirchhofer D. Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists. Nat Struct Mol Biol. 2017 Aug 21. doi: 10.1038/nsmb.3453. PMID:28825733 doi:http://dx.doi.org/10.1038/nsmb.3453
- ↑ Giunzioni I, Tavori H. New developments in atherosclerosis: clinical potential of PCSK9 inhibition. Vasc Health Risk Manag. 2015 Aug 24;11:493-501. doi: 10.2147/VHRM.S74692., eCollection 2015. PMID:26345307 doi:http://dx.doi.org/10.2147/VHRM.S74692
- ↑ Mitchell T, Chao G, Sitkoff D, Lo F, Monshizadegan H, Meyers D, Low S, Russo K, DiBella R, Denhez F, Gao M, Myers J, Duke G, Witmer M, Miao B, Ho SP, Khan J, Parker RA. Pharmacologic Profile of the Adnectin BMS-962476, a Small Protein Biologic Alternative to PCSK9 Antibodies for LDL Lowering. J Pharmacol Exp Ther. 2014 Jun 10. pii: jpet.114.214221. PMID:24917546 doi:http://dx.doi.org/10.1124/jpet.114.214221
- ↑ Mullard A. Nine paths to PCSK9 inhibition. Nat Rev Drug Discov. 2017 Apr 28;16(5):299-301. doi: 10.1038/nrd.2017.83. PMID:28450722 doi:http://dx.doi.org/10.1038/nrd.2017.83