Journal:Genes:1
From Proteopedia
(Difference between revisions)

| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene='77/775919/Cv/ | + | <StructureSection load='' size='450' side='right' scene='77/775919/Cv/10' caption=''> |
=== A DNA Structural Alphabet Distinguishes Structural Features of DNA Bound to Regulatory Proteins and in the Nucleosome Core Particle === | === A DNA Structural Alphabet Distinguishes Structural Features of DNA Bound to Regulatory Proteins and in the Nucleosome Core Particle === | ||
<big>Bohdan Schneider, Paulina Bozikova, Petr Cech, Daniel Svozil and Jiri Cerny</big> <ref>doi 10.3390/genes8100278</ref> | <big>Bohdan Schneider, Paulina Bozikova, Petr Cech, Daniel Svozil and Jiri Cerny</big> <ref>doi 10.3390/genes8100278</ref> | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
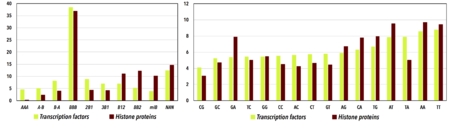
| - | We observe significant structural differences between DNA in complexes with transcription factors and with histone proteins. To analyze these two types of structures, we used the DNA structural alphabet called CANA, which we had developed earlier by analysis of hundreds of crystal structures containing DNA molecules <ref> | + | We observe significant structural differences between DNA in complexes with transcription factors and with histone proteins. To analyze these two types of structures, we used the DNA structural alphabet called CANA, which we had developed earlier by analysis of hundreds of crystal structures containing DNA molecules ([[Journal:Acta_Cryst_D:2]]) <ref>doi 10.1107/S2059798318000050</ref>. The structural alphabet allows to "translate" a three-dimensional (spatial, 3D) structure of DNA building blocks called dinucleotides into a series of letters. Each letter then represents the structure of a dinucleotide and a chain of these letters can be analyzed as a "word" representing the whole 3D DNA structure. The process of translation of a 3D structure to the alphabet letters can be performed at the web server [https://dnatco.org dnatco.org] <ref>pmid 27150812</ref>. |
Different patterns of the CANA letters observed in complexes with transcription factors and with histone proteins can be interpreted as features discriminating the specific of DNA to transcription factors from non-specific binding to histone proteins. Especially noteworthy is the role of two DNA structural forms, so called A-DNA and BII-DNA ("B-two-DNA"), which are described by the CANA letters AAA and BB2. The AAA structures are found quite frequently in DNA specifically bound to transcription factors at regions where the DNA duplex bends around the protein. AAA is avoided in non-specific complexes with histone proteins, where BB2 plays the essential role: the wrapping of the DNA duplex around the histone proteins can be explained by the periodic occurrence of the CANA letter BB2 every 10.3 steps along the DNA strand. | Different patterns of the CANA letters observed in complexes with transcription factors and with histone proteins can be interpreted as features discriminating the specific of DNA to transcription factors from non-specific binding to histone proteins. Especially noteworthy is the role of two DNA structural forms, so called A-DNA and BII-DNA ("B-two-DNA"), which are described by the CANA letters AAA and BB2. The AAA structures are found quite frequently in DNA specifically bound to transcription factors at regions where the DNA duplex bends around the protein. AAA is avoided in non-specific complexes with histone proteins, where BB2 plays the essential role: the wrapping of the DNA duplex around the histone proteins can be explained by the periodic occurrence of the CANA letter BB2 every 10.3 steps along the DNA strand. | ||
| Line 15: | Line 15: | ||
{{Clear}} | {{Clear}} | ||
| - | *<scene name='77/775919/Cv/ | + | *<scene name='77/775919/Cv/11'>The duplex</scene> displays DNA from the structure of human TFIIB-related factor 2 and TATA box binding protein bound to U6#2 promoter DNA (PDB code [[4roc]]), where the DNA backbone acquires the A-DNA form (CANA letter AAA) at the bend. Dinucleotides adopting the structure described by the CANA letter AAA (BB2) are highlighted in red (blue) color. |
| - | *<scene name='77/775919/Cv/ | + | *<scene name='77/775919/Cv/10'>The duplex</scene> depicts first 75 base pairs from DNA in NCP of the PDB code [[5f99]]. |
</StructureSection> | </StructureSection> | ||
<references/> | <references/> | ||
__NOEDITSECTION__ | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
- ↑ Schneider B, Bozikova P, Cech P, Svozil D, Cerny J. A DNA Structural Alphabet Distinguishes Structural Features of DNA Bound to Regulatory Proteins and in the Nucleosome Core Particle. Genes (Basel). 2017 Oct 18;8(10). pii: genes8100278. doi: 10.3390/genes8100278. PMID:29057824 doi:http://dx.doi.org/10.3390/genes8100278
- ↑ Bohdan Schneider, Paulina Bozikova, Iva Necasova, Petr Cech, Daniel Svozilb and Jirı Cerný. A DNA structural alphabet provides new insight into DNA flexibility. Acta Cryst. (2018). D74, 52-64 doi:http://dx.doi.org/10.1107/S2059798318000050
- ↑ Cerny J, Bozikova P, Schneider B. DNATCO: assignment of DNA conformers at dnatco.org. Nucleic Acids Res. 2016 Jul 8;44(W1):W284-7. doi: 10.1093/nar/gkw381. Epub 2016, May 5. PMID:27150812 doi:http://dx.doi.org/10.1093/nar/gkw381
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.