|
|
| (2 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| | | | |
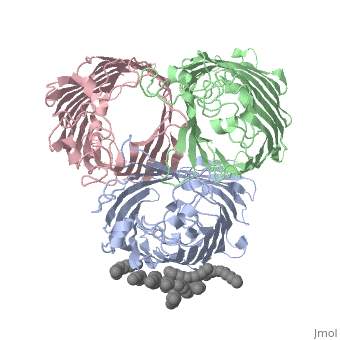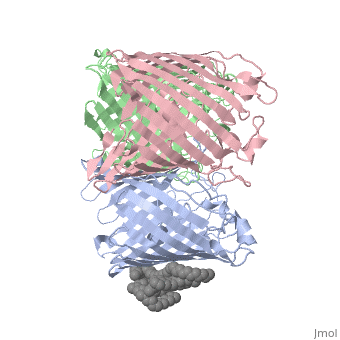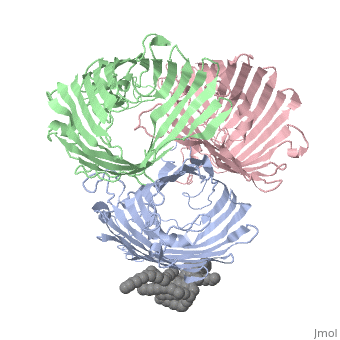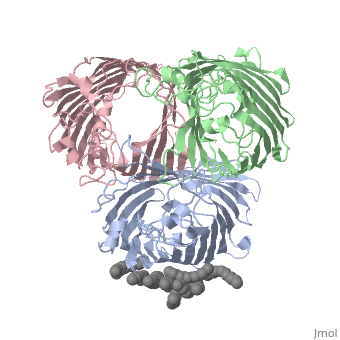
| | ==OSMOPORIN (OMPK36) FROM KLEBSIELLA PNEUMONIAE== | | ==OSMOPORIN (OMPK36) FROM KLEBSIELLA PNEUMONIAE== |
| - | <StructureSection load='1osm' size='340' side='right' caption='[[1osm]], [[Resolution|resolution]] 3.20Å' scene=''> | + | <StructureSection load='1osm' size='340' side='right'caption='[[1osm]], [[Resolution|resolution]] 3.20Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[1osm]] is a 3 chain structure with sequence from [http://en.wikipedia.org/wiki/"bacillus_pneumoniae"_(schroeter_1886)_flugge_1886 "bacillus pneumoniae" (schroeter 1886) flugge 1886]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1OSM OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1OSM FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[1osm]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Klebsiella_pneumoniae Klebsiella pneumoniae]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1OSM OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1OSM FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=D12:DODECANE'>D12</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.2Å</td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1osm FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1osm OCA], [http://pdbe.org/1osm PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1osm RCSB], [http://www.ebi.ac.uk/pdbsum/1osm PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=1osm ProSAT]</span></td></tr> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=D12:DODECANE'>D12</scene></td></tr> |
| | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1osm FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1osm OCA], [https://pdbe.org/1osm PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1osm RCSB], [https://www.ebi.ac.uk/pdbsum/1osm PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1osm ProSAT]</span></td></tr> |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[http://www.uniprot.org/uniprot/OMPC_KLEPN OMPC_KLEPN]] Forms pores that allow passive diffusion of small molecules across the outer membrane. In K.pneumoniae it has been shown to bind the C1Q component and activate the classical pathway of the complement system. | + | [https://www.uniprot.org/uniprot/OMPC_KLEPN OMPC_KLEPN] Forms pores that allow passive diffusion of small molecules across the outer membrane. In K.pneumoniae it has been shown to bind the C1Q component and activate the classical pathway of the complement system. |
| | == Evolutionary Conservation == | | == Evolutionary Conservation == |
| | [[Image:Consurf_key_small.gif|200px|right]] | | [[Image:Consurf_key_small.gif|200px|right]] |
| Line 30: |
Line 31: |
| | | | |
| | ==See Also== | | ==See Also== |
| - | *[[Osmoporin OmpK36 (K. pneumoniae)|Osmoporin OmpK36 (K. pneumoniae)]] | + | *[[Porin 3D structures|Porin 3D structures]] |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Dutzler, R]] | + | [[Category: Klebsiella pneumoniae]] |
| - | [[Category: Schirmer, T]] | + | [[Category: Large Structures]] |
| - | [[Category: Beta-barrel]] | + | [[Category: Dutzler R]] |
| - | [[Category: Non-specific porin]] | + | [[Category: Schirmer T]] |
| - | [[Category: Osmoporin]]
| + | |
| - | [[Category: Outer membrane protein]]
| + | |
| - | [[Category: Transmembrane]]
| + | |
| Structural highlights
Function
OMPC_KLEPN Forms pores that allow passive diffusion of small molecules across the outer membrane. In K.pneumoniae it has been shown to bind the C1Q component and activate the classical pathway of the complement system.
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
BACKGROUND: Porins are channel-forming membrane proteins that confer solute permeability to the outer membrane of Gram-negative bacteria. In Escherichia coli, major nonspecific porins are matrix porin (OmpF) and osmoporin (OmpC), which show high sequence homology. In response to high osmolarity of the medium, OmpC is expressed at the expense of OmpF porin. Here, we study osmoporin of the pathogenic Klebsiella pneumoniae (OmpK36), which shares 87% sequence identity with E. coliOmpC in an attempt to establish why osmoporin is best suited to function at high osmotic pressure. RESULTS: The crystal structure of OmpK36 has been determined to a resolution of 3.2 A by molecular replacement with the model of OmpF. The structure of OmpK36 closely resembles that of the search model. The homotrimeric structure is composed of three hollow 16-stranded antiparallel beta barrels, each delimiting a separate pore. Most insertions and deletions with respect to OmpF are found in the loops that protrude towards the cell exterior. A characteristic ten-residue insertion in loop 4 contributes to the subunit interface. At the pore constriction, the replacement of an alanine by a tyrosine residue does not alter the pore profile of OmpK36 in comparison with OmpF because of the different course of the mainchain. Functionally, as characterized in lipid bilayers and liposomes, OmpK36 resembles OmpC with decreased conductance and increased cation selectivity in comparison with OmpF. CONCLUSIONS: The osmoporin structure suggests that not an altered pore size but an increase in charge density is the basis for the distinct physico-chemical properties of this porin that are relevant for its preferential expression at high osmotic strength.
Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae.,Dutzler R, Rummel G, Alberti S, Hernandez-Alles S, Phale P, Rosenbusch J, Benedi V, Schirmer T Structure. 1999 Apr 15;7(4):425-34. PMID:10196126[1]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Dutzler R, Rummel G, Alberti S, Hernandez-Alles S, Phale P, Rosenbusch J, Benedi V, Schirmer T. Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae. Structure. 1999 Apr 15;7(4):425-34. PMID:10196126
|