1shy
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
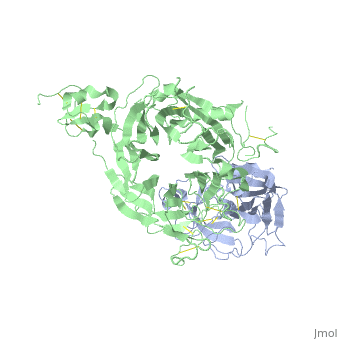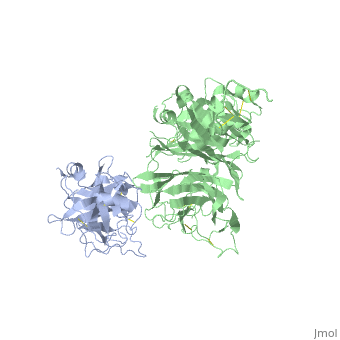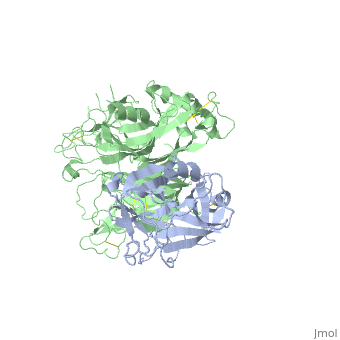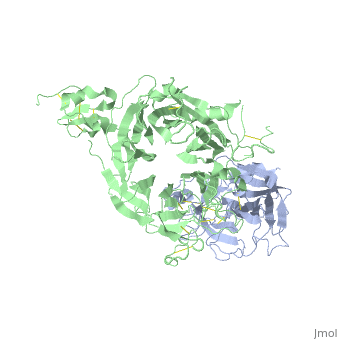
==The Crystal Structure of HGF beta-chain in Complex with the Sema Domain of the Met Receptor.== | ==The Crystal Structure of HGF beta-chain in Complex with the Sema Domain of the Met Receptor.== | ||
| - | <StructureSection load='1shy' size='340' side='right' caption='[[1shy]], [[Resolution|resolution]] 3.22Å' scene=''> | + | <StructureSection load='1shy' size='340' side='right'caption='[[1shy]], [[Resolution|resolution]] 3.22Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1shy]] is a 2 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1shy]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1SHY OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1SHY FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.22Å</td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1shy FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1shy OCA], [https://pdbe.org/1shy PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1shy RCSB], [https://www.ebi.ac.uk/pdbsum/1shy PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1shy ProSAT]</span></td></tr> |
</table> | </table> | ||
== Disease == | == Disease == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/HGF_HUMAN HGF_HUMAN] Defects in HGF are the cause of deafness autosomal recessive type 39 (DFNB39) [MIM:[https://omim.org/entry/608265 608265]. A form of profound prelingual sensorineural hearing loss. Sensorineural deafness results from damage to the neural receptors of the inner ear, the nerve pathways to the brain, or the area of the brain that receives sound information.<ref>PMID:19576567</ref> |
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/HGF_HUMAN HGF_HUMAN] Potent mitogen for mature parenchymal hepatocyte cells, seems to be a hepatotrophic factor, and acts as a growth factor for a broad spectrum of tissues and cell types. Activating ligand for the receptor tyrosine kinase MET by binding to it and promoting its dimerization.<ref>PMID:15167892</ref> <ref>PMID:20624990</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 21: | Line 21: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1shy ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1shy ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The Met tyrosine kinase receptor and its ligand, hepatocyte growth factor (HGF), play important roles in normal development and in tumor growth and metastasis. HGF-dependent signaling requires proteolysis from an inactive single-chain precursor into an active alpha/beta-heterodimer. We show that the serine protease-like HGF beta-chain alone binds Met, and report its crystal structure in complex with the Sema and PSI domain of the Met receptor. The Met Sema domain folds into a seven-bladed beta-propeller, where the bottom face of blades 2 and 3 binds to the HGF beta-chain 'active site region'. Mutation of HGF residues in the area that constitutes the active site region in related serine proteases significantly impairs HGF beta binding to Met. Key binding loops in this interface undergo conformational rearrangements upon maturation and explain the necessity of proteolytic cleavage for proper HGF signaling. A crystallographic dimer interface between two HGF beta-chains brings two HGF beta:Met complexes together, suggesting a possible mechanism of Met receptor dimerization and activation by HGF. | ||
| - | |||
| - | Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor.,Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C EMBO J. 2004 Jun 16;23(12):2325-35. Epub 2004 May 27. PMID:15167892<ref>PMID:15167892</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1shy" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[Hepatocyte growth factor|Hepatocyte growth factor]] | *[[Hepatocyte growth factor|Hepatocyte growth factor]] | ||
| - | *[[Hepatocyte growth factor receptor|Hepatocyte growth factor receptor]] | + | *[[Hepatocyte growth factor receptor 3D structures|Hepatocyte growth factor receptor 3D structures]] |
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Homo sapiens]] |
| - | [[Category: | + | [[Category: Large Structures]] |
| - | [[Category: | + | [[Category: Stamos J]] |
| - | [[Category: | + | [[Category: Wiesmann C]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
The Crystal Structure of HGF beta-chain in Complex with the Sema Domain of the Met Receptor.
| |||||||||||