User:Karsten Theis/RNaseA physical model explanation
From Proteopedia
< User:Karsten Theis(Difference between revisions)
(New page: ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene=''> == Function == == Disease == ==...) |
(→Introduction) |
||
| (23 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | ==Introduction== |
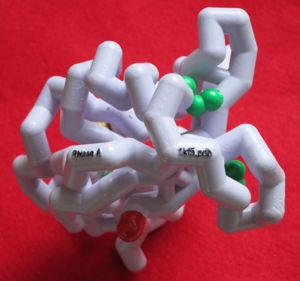
| - | <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene=''> | + | This proteopedia page is intended as a companion to a physical model of RNase A used for teaching protein folding and structure at Westfield State University. |
| + | |||
| + | [[Image:RNAseA model.jpg|300px]] | ||
| + | |||
| + | |||
| + | Mark Hoelzer from the MSOE Center for Biomolecular Modeling [https://www.centerforbiomolecularmodeling.org/about] designed, printed and painted the model. Having the model in your hand is great because you can experience the structure in a very direct way, and having this companion page is great because you can see the model even if you do not have the physical model in your hand. It also allows you to hover over different parts of the model with a mouse to get more information (try it after turning off spinning with the +/- spin button!). | ||
| + | |||
| + | ==Tour of the structure== | ||
| + | <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene='78/785360/Physmodel/1'> | ||
| + | |||
| + | ==What the model shows== | ||
| + | |||
| + | ===Trace of alpha carbon atoms=== | ||
| + | <jmol><jmolLink> | ||
| + | <script> select (*.CB and not *%B) or (GLY.CA and not *%B); label "%r"; color label black; | ||
| + | </script> | ||
| + | <text>label residues</text> | ||
| + | </jmolLink></jmol> (click +/- labels to turn off again) | ||
| + | |||
| + | |||
| + | ===Hydrogen bonds=== | ||
| + | <jmol><jmolLink> | ||
| + | <script> select all; color hbonds blue; delay 0.8; color hbonds [xCFd0FF] | ||
| + | </script> | ||
| + | <text>highlight hydrogen bonds</text> | ||
| + | </jmolLink></jmol> | ||
| + | |||
| + | ===N-terminus and C-terminus=== | ||
| + | |||
| + | <jmol><jmolLink> | ||
| + | <script> select 1.CA; spacefill 200%; delay 0.8; spacefill off; | ||
| + | </script> | ||
| + | <text>N-terminus</text> | ||
| + | </jmolLink></jmol> and <jmol><jmolLink> | ||
| + | <script> select 124.CA; spacefill 200%; delay 0.8; spacefill off; | ||
| + | </script> | ||
| + | <text>C-terminus</text> | ||
| + | </jmolLink></jmol> refer to... | ||
| + | |||
| + | ===Disulfide bridges=== | ||
| + | |||
| + | <jmol><jmolLink> | ||
| + | <script> select sidechain and Cys and not Cys%B; spacefill 150%; delay 0.8; spacefill 75%; | ||
| + | </script> | ||
| + | <text>Disulfide bridges</text> | ||
| + | </jmolLink></jmol> are ... | ||
| + | |||
| + | ==Tour of things not shown in the physical model== | ||
| + | |||
| + | ===Substrate binding=== | ||
| + | <scene name='78/785360/Substrate/1'> | ||
| + | Substrate binding</scene> | ||
| + | |||
| + | ===Shape of the protein=== | ||
| + | <jmol><jmolLink> | ||
| + | <script> select protein; isosurface ignore (not selected) SASURFACE 1.2 | ||
| + | </script> | ||
| + | <text>molecular surface</text> | ||
| + | </jmolLink></jmol> | ||
| + | |||
| + | ===Active site residues=== | ||
| + | <scene name='78/785360/Acidbase/1'>Active site residues</scene> | ||
| + | |||
| + | ===Hydrophobic core=== | ||
| + | <scene name='78/785360/Core/1'>Hydrophobic core</scene> | ||
| + | |||
| + | |||
| + | <scene name='78/785360/Physmodel/1'>Back to overall view</scene> | ||
== Function == | == Function == | ||
Current revision
Introduction
This proteopedia page is intended as a companion to a physical model of RNase A used for teaching protein folding and structure at Westfield State University.
Mark Hoelzer from the MSOE Center for Biomolecular Modeling [1] designed, printed and painted the model. Having the model in your hand is great because you can experience the structure in a very direct way, and having this companion page is great because you can see the model even if you do not have the physical model in your hand. It also allows you to hover over different parts of the model with a mouse to get more information (try it after turning off spinning with the +/- spin button!).
Tour of the structure
| |||||||||||