|
|
| (18 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | __TOC__
| + | =Histone Lysine Methyltransferase: Gene Activator= |
| - | === Overview ===
| + | |
| | | | |
| - | The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM)<ref name="Corbo2013">PMID:23685143</ref>.
| + | <StructureSection load='1o9s' size='350' frame='true' side='right' caption='Lysine Methyl Transferase' scene='81/811092/Kmt_full/2'> |
| | | | |
| - | <StructureSection load='2i2y' size='400' frame='true' side='right' caption='SRp20 bound to RNA ligand (PDB entry [[2i2y]])' scene='78/782597/Rrmredgreen/2'>
| + | |
| - | == Introduction ==
| + | |
| - | === History ===
| + | |
| - | Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins ([https://en.wikipedia.org/wiki/Heterogeneous_ribonucleoprotein_particle hnRNPs]).
| + | |
| - | The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus<ref name="Zahler">PMID:1577277</ref>. The discovery of this family started in the 1900s with the [https://en.wikipedia.org/wiki/Serine/arginine-rich_splicing_factor_1 SF2] (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight<ref name="Zahler">PMID:1577277</ref>.
| + | |
| - | An identical protein, called [http://www.uniprot.org/uniprot/Q9V3V0 X16], was discovered in an earlier paper studying different genes that change expression during [https://en.wikipedia.org/wiki/B_cell B-cell] development<ref name="Corbo2013">PMID:23685143</ref>. At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true<ref name="Ayane">PMID:2030943</ref><ref name="Cacero">PMID:11932019</ref>.
| + | |
| - | The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease<ref name="Corbo2013">PMID:23685143</ref>. SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4<ref name="Corbo2013">PMID:23685143</ref>. Another function of SRp20 is its role in export of mRNA out of the nucleus, notably [https://en.wikipedia.org/wiki/Histone_H2A H2A histone] mRNA export<ref name="Hargous">PMID:17036044</ref>.
| + | |
| - | == Structure and Function ==
| + | |
| - | === Structure Determination ===
| + | |
| | | | |
| - | Attempts to determine the structure of native SRp20 have been largely unsuccessful due to the low solubility of the protein. This is likely due to the hydrophobic core of the RRM and exposed hydrophobic residues for RNA recognition on the β-sheets. As a solution, researchers removed the SR domain from the C terminus, leaving only the SRp20 RRM and a small arginine rich segment at the C terminus, then fused with a soluble <scene name='78/782597/Imager0/2'>IgG binding domain</scene> of Streptococcal protein G to the N terminus of the protein, providing the first published structure of the SRp20 RRM via NMR. However, the solution of the structure via [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR], in addition to fusion with a globular tag, results in multiple possible conformations of the protein, meaning measurements such as bond angles, lengths, and substrate interactions are variable. Further, information concerning structural aspects of the SR domain are still limited to experimental data of protein function with certain mutations or deletions, and by comparison to sister proteins such as 9G8. To date, structure of the SR domain or the protein without the globular tag have not been solved, nor has a crystal structure for any part of the protein been determined<ref name="Hargous">PMID:17036044</ref>.
| + | == Introduction == |
| | | | |
| - | == Splicing Activity ==
| + | |
| - | The splicing mechanism for SRp20 follows the normal eukaryotic mechanism, in which five different [https://en.wikipedia.org/wiki/SnRNP small nuclear ribonucleoproteins] (snRNPs) bring the splice sites together in order to start the reaction (Figure 1). Specifically, SRp20 and other SR proteins interact with the RNA ligand at the [https://en.wikipedia.org/wiki/Exonic_splicing_enhancer exonic splicing enhancer sequence] at the beginning of the 3’ splice site adjacent to the intron being removed. SRp20 facilitates the interaction of the U2 snRNP with the RNA to continue the mechanism (Figure 2)<ref name="Shepard">PMID:19857271</ref>. [[Image:mechanism1.png|300px|left|thumb|'''Figure 1.''' Splicing mechanism for eukaryotes. Free 3’OH nucleophile of adenosine in intron attacks phosphorus of phosphate creating a ring structure intron known as a lariat. The free 2’OH in the 5’ splice site (red) acts as the nucleophile to attack the phosphate of the first nucleotide in the 3’ splice site (red) to release the [https://news.brown.edu/articles/2012/06/lariats lariat] intron. The products include the final modified RNA sequence and the lariat which will be recycled.]] [[Image:prettymechanism.png|260px|right|thumb|'''Figure 2.''' SRp20 works with [https://en.wikipedia.org/wiki/U2_spliceosomal_RNA U2] snRNP: Five snRNPs are needed in the eukaryotic splicing mechanism to facilitate the reaction. The U2 snRNP must attach to the 3’ splice site to bring together the 5’ and 3’ splice sites (red). SRp20 facilitates binding of U2 to the 3’ splice site by binding to the exonic splicing enhancer sequence (blue) in the RNA at the backbone. U2 must bind before the other snRNPs can bind to continue the mechanism.]]
| + | |
| | | | |
| | + | ===Histone Methylation=== |
| | | | |
| - | == Structural Highlights ==
| + | [[Image:human_nucleosome_ray_trace.png|200 px| right| thumb|Human nucleosome particle, pbd code: 5y0c]] |
| | | | |
| - | === RNA Recognition Motif ===
| + | [https://en.wikipedia.org/wiki/Histone Histone proteins] aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer of core proteins H2a, H2b, H3, and H4, which organize into two heterodimers; H1 and H5 act as linker proteins. About 145-157 base pairs wind around a histone heterodimer core. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of transcription factors in the genome, either promoting or inhibiting transcription. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> |
| - | The SRP20 RRM (aa 1-86) a βαββαβ <scene name='78/782597/Imager1/3'>pattern</scene>, common of many other RRMs3. For substrate binding, researchers used a 4 base RNA with sequence CAUC, which matches the SRP20 recognition sequence found on corresponding H2A mRNA. The RNA bases each <scene name='78/782597/Imager2/5'>stack</scene> onto an aromatic side chain protruding from one of the SRP20 β-sheets, forming the primary interactions which allow substrate binding to the protein. In particular, C1 <scene name='78/782597/Imager3/2'>stacks</scene> on Y13 in β1, <scene name='78/782597/Imager4/2'>A2</scene> stacks on F50 in β3, and F48 of β3 sits in between the sugar rings of C1 and A2. It should also be noted that A2 adopts an irregular <scene name='78/782597/Imager5/3'>syn</scene> conformation when bound to the RRM, something that was observed only for guanine in the 2 position previously. U3 <scene name='78/782597/Imager8/4'>Stacks</scene> onto F48 in β3, also W40 and A42 in β2, however when bound, U3 <scene name='78/782597/Imager9/2'>bulges</scene> out of line in comparison to the rest of the substrate. C4 partially stacks over <scene name='78/782597/Imager6/4'>A2</scene>, and also forms hydrogen <scene name='78/782597/Imager7/3'>bonds</scene> between the C4 amino group, A2 2’ oxygen, and a main chain phosphate oxygen.
| + | |
| - | While all 4 bases form a number of hydrophobic stacking interactions, alteration to the last 3 bases of substrate sequence does not significantly impact binding affinity, while C to G mutation of C1 results in a 10-fold decrease in binding affinity. This suggests that C1 interacts specifically with the protein, while positions 2-4 interact nonspecifically3. The Srp20 RRM is able to recognize C1 with high specificity primarily through 4 <scene name='78/782597/Imager10/4'>hydrogen bonds</scene>: from the C1 amino protons to Leu 80 backbone carbonyl oxygen and to Glu 79 side-chain carbonyl oxygen, from C1 N3 to Asn82 amide, and C1 O2 with Ser 81 side chain hydroxyl group.
| + | |
| - | The semi specific RNA recognition is a mechanism which reduces evolutionary pressure on bound mRNA by increasing the number of possible RNA recognition sequences. As a result, tolerance for possible mutation in the RNA sequence is increased, meaning Srp20 can bind a more diverse range of substrates, or even original substrates that were mutated during replication (eg. H2A mRNA with a point mutation) thereby increasing organism survival chance by reducing the probability of physiological impact as a result of certain mutations<ref name="Hargous">PMID:17036044</ref>.
| + | |
| | | | |
| - | === Tip Associated Protein Binding Domain === | + | Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_domain/2'>SET7 domain</scene> family, which can target H3, H4, or H2a. Sites known for gene activation are Lysine-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys- 27 on H3 and Lys-20 on H4 are known for gene inactivation.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity; specifics of this characteristic depend on site and species of organism. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as [https://www.ncbi.nlm.nih.gov/books/NBK22268/ p53] are site specifically methylated to promote biological function <ref name = "Rizzo" />, whereas hypomethylation of [https://en.wikipedia.org/wiki/CpG_site#CpG_islands CpG] is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />, <ref name="Dong" /> |
| - | [[Image:AASequence.jpg|350px|right|thumb|'''Figure 3.''' SRp20 Domain Representation. Shown are the RRM (green), the TAP binding linker (red line) and SR-rich domain (blue).]]
| + | |
| - | In addition to RNA recognition and alternative splicing functions, SRP20 has been shown to associate with Tip Associated Protein (TAP), an mRNA export factor, to promote transport of bound mRNA out of the nucleus for eventual translation3. In particular SRp20 promotes the export of H2A histone mRNA, by binding the CAUC consensus sequence on the mRNA and binding TAP. Previous experiments have shown that Srp20 binding TAP is dependent on presence of both the <scene name='78/782597/Rrmredgreen/1'>SRp20 RRM</scene> and a short arginine rich C-terminal segment after the RRM (aa 1-83 and 84-90 respectively) ('''Figure 3'''). Previous research also shows that mutation of any one of the three arginine residues present between residues 84-90 to glutamate prevents TAP binding, indicative of the importance of these arginine residues in TAP association. The structure is only solved to residue 86 so only the <scene name='78/782597/Arginine1/4'>first arginine residue</scene> is present. The same study also found that transfer of the TAP-binding motif to a non-functional REF2 RRM still allowed for TAP-binding and nuclear export of the target protein, suggesting that not only is the TAP-binding motif transferable, it does not depend on interaction with the host RRM to retain function<ref name="Hargous">PMID:17036044</ref>.
| + | |
| | | | |
| - | === SR Domain ===
| + | |
| - | While the RNA-protein interaction occurs at the RRMs, SR domains are typically responsible for the recruitment of other proteins that act in the splicing mechanism. The serines within the SR domain are phosphorylated by kinases within the cell to direct them to pre-mRNA sites (Figure 4). [[Image:phosphorylatedserine.png|300px|right|thumb|'''Figure 4.''' An example of one phosphorylated serine within the RS domain.]] Phosphorylation acts as recruiting tools for the SRp20 protein and other SR proteins to promote splicing. However, some SR proteins have shown that phosphorylation actually leads to a decrease in splicing, as in the [https://www.ncbi.nlm.nih.gov/pubmed/14765198 SRp38] protein undergoing heat shock. Research has recently revealed that serines are dephosphorylated as splicing continues, indicating how far along the mRNA strand is in splicing. Dephosphorylation then serves as a signal to the cell that the mRNA is ready to be exported out of the nucleus. Rephosphorylation then appears to trigger the SR protein to enter back into the nucleus.
| + | |
| - | The SR domain also appears to modulate mRNA stability, though different SR proteins have been shown to have greater effects. Some SR proteins appears to increase the sensitivity of certain mRNA sequences to degradation; this degradation appears to be controlled by SR domain interactions with the 3’ UTR of the pre-mRNA. These effects are not present without the SR domain of the protein. The destabilizing effect of SRp20 specifically has yet to be studied<ref name="Hargous">PMID:17036044</ref>.
| + | |
| | | | |
| - | == 9G8 and SRP20 == | + | ==Lysine Methyltransferase (KMT) Structure== |
| - | [https://en.wikipedia.org/wiki/SFRS7 9G8] is another SR protein that is 80% similar in amino acid sequence. They are two of the smallest proteins in the SR family and both contain an RRM that promotes export of mRNA through interaction with the TAP protein. The only other protein shown to promote transport through the TAP protein is SF2, one of the first SR proteins discovered<ref name="Huang">PMID:12667464</ref>. Both RRMs adopt a βαββαβ mentioned earlier. Both RRMs interact with RNA with limited selectivity and therefore recognize many different RNA sequences. The 9G8 RRM contains an large hydrophobic core on its B-sheet. Observing the 9G8 protein has proved useful in understanding SRp20 less stable protein structure. In fact, the one of the only significant structural difference between the two proteins lies in the loops between a-helix 2 and B-4 where several amino acids are not conserved.
| + | |
| - | Aside from the RRM, both proteins have one SR-rich domain although 9G8 includes about 40 more amino acids this domain<ref name="Corbo2013">PMID:23685143</ref>. [[Image:9G8srp20comparison.png|300px|right|thumb|'''Figure 5.''' SRp20 and 9G8 proteins. The PDB file for SRp20 (left; PDB file: 2i2y) does not include the SR-rich domain but 9G8 (right; PDB file: 2hvz) does. Boxed image shows an RSR region in the PG8 protein with oxygens highlighted in red. Images taken from PyMol software.jpg]] Within the bigger 9G8 protein, there is a zinc knuckle that allows for binding of pyrimidine-rich RNA sequences. This zinc knuckle is not present in SRp20, lending the protein to binding of more purine-rich sequences<ref name="Huang">PMID:12667464</ref>. Not only are these two proteins similar, but they also play similar roles in mRNA export out of the nucleus. Both move continuously between the nucleus and cytoplasm which requires phosphorylation of its serine residues located in the SR-rich domain. Serine phosphorylation has been shown to have great importance in the proper functioning of many SR proteins (Figure 4)<ref name="Shepard">PMID:19857271</ref>.
| + | |
| - | </StructureSection>
| + | |
| - | == Medical Significance ==
| + | |
| - | === Cancer ===
| + | |
| - | SRp20 has been linked to [https://en.wikipedia.org/wiki/Cancer cancer] in many instances, as well as other AS proteins. SRp20 has been seen to activate the AS of the [http://proteopedia.org/wiki/index.php/CD44 CD44 adhesion molecule.] SRp20 facilitates the splicing of exon v9, one important for function of CD44. Loss of function of SRp20 leading to loss of function of CD44 will lead to loss of “stickiness” of cells, a way that cancer cells can spread to other areas of the body. SRp20 can also affect the alternative splicing of [https://en.wikipedia.org/wiki/Oncogene oncogenes] and [https://en.wikipedia.org/wiki/Tumor_suppressor_gene tumor suppressors]. Expression of the signaling pathway for SRp20 translation is directly triggered by oncogenic signalling pathways. For example, the [https://en.wikipedia.org/wiki/Wnt_signaling_pathway Wnt] pathway that is associated with cancer development also drives further expression of SRp20 and other SR proteins<ref name="Corbo2012">PMID:22623141</ref>. These proteins aid in cell growth and proliferation, in that they increase expression of proteins they are involved in splicing. One study found that silencing SRp20 slowed cell proliferation, further supporting the idea that SRp20 helps with this process<ref name="Corbo2013">PMID:23685143</ref>. Cell cycle regulator proteins [http://proteopedia.org/wiki/index.php/FOXM1 FoxM1], [https://en.wikipedia.org/wiki/CDC25B Cdc25B], and [http://proteopedia.org/wiki/index.php/Plk1 PLK1] are alternatively spliced by SRp20, specifically regulating the G2/M cycles. These proteins are also associated with regulation of cell apoptosis. When these genes are overexpressed due to overexpressed SRp20, cells will no longer be able to apoptose when they are damaged, and the cell cycle will continue without regulation, causing overactive cell proliferation and eventually cancer. SRp20 has been seen to be overexpressed in ovarian, lung, breast, stomach, skin, bladder, colon, liver, thyroid, and kidney cancer tissue<ref name="Jia">PMID:21179588</ref>. SRp20 also regulates genes associated with cellular senescence, or cellular immortality, which contributes to cancer formation. Specifically, SRp20 alternatively splices the [https://ghr.nlm.nih.gov/gene/TP53 TP53] gene, which generates the [http://proteopedia.org/wiki/index.php/P53 p53] senescence protein.
| + | |
| | | | |
| - | === Neurological Disorders ===
| + | The structure of human histone methyltransferase SET7/9 was crystallized using x-ray diffraction at 1.75Å in its product form <ref name="Xiao" />. SET7/9 was crystallized with its cofactor was in its unmethylated form, S-adenosyl-Lhomocysteine (SAH), and with its product, a ten residue peptide with a methylated lysine at residue 4. |
| - | Along with cancer, SRp20 mutations have been linked to [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimers], a neurodegenerative disorder. SRp20 is involved in AS of a wide array of RNAs, including that of the [https://en.wikipedia.org/wiki/Tropomyosin_receptor_kinase_B TRKB] gene to generate TrkB-Shc transcripts that are involved in generation of the disorder. SRp20 also promotes exclusion of exon 10 in the [http://proteopedia.org/wiki/index.php/Tau TAU] gene, a gene important in establishing microtubules in axons or transport processes.Underexpression of SRp20 results in dysfunction of the TAU gene, less microtubule functionality, and the brain deterioration characteristic of Alzheimers<ref name="Ebneth">PMID:9813097</ref><ref name="Corbo2012">PMID:22623141</ref>.
| + | |
| | | | |
| - | === Other Genetic Disorders ===
| + | |
| - | SRp20 and other SR proteins have been shown to prevent [https://en.wikipedia.org/wiki/R-loop R-loops] from forming, 3-stranded nucleic acid structures consisting of RNA and DNA. R-loops have been known to promote mutations, recombination, and chromosome rearrangement. One proposed mechanism for R-loop prevention by SRp20 is that SRp20, being a protein involved in RNA metabolism, is a binding partner of the [https://www.uniprot.org/uniprot/P11387 TOP1] protein. Underexpression of TOP1 also promotes R-loop formation. TOP1 has kinase activity that potentially phosphorylates the SR domain of SRp20, which contributes to its function. Underexpression of TOP1 would lead to loss-of-function of SRp20, which could lead to cancer and Alzheimers as mentioned above, as well as cause R-loops to form. R-loops have been associated with disorders such as [https://www.ndss.org/about-down-syndrome/down-syndrome/ Down Syndrome]<ref name="Naro">PMID:25926848</ref>.
| + | |
| | | | |
| - | == References == | + | ===Overall Secondary Structure=== |
| - | <references/> | + | |
| | + | Due to the composition of its secondary structure, KMT is an alpha-beta protein <ref name="Xiao" />. The helical composition includes 3 <scene name='81/811086/Alpha_helices/4'>alpha helices</scene>, with two residing in the SET domain and one in the C-terminal domain. The alpha helices in the SET domain are two turns while the C-terminal helix is by far the largest with 4 turns. There are also 2 <scene name='81/811086/3-10_helices/4'>3-10 helices</scene> in the SET domain which are each one turn. There are 21 total <scene name='81/811086/Beta_sheets/3'>beta strands</scene> which reside in the N-terminal domain and the SET domain. The beta strands are primarily anti-parallel and multiple antiparallel strands are connected by Type 1 and Type 2 <scene name='81/811086/Beta_turns/3'>beta turns</scene>. |
| | + | |
| | + | |
| | + | |
| | + | ===The Active Site=== |
| | + | |
| | + | The active site and binding pocket of KMT contain residues and shape that optimize catalytic function and stability. First, the lysine of the histone enters the active site via the <scene name='81/811092/Tyrosine_channel_2/3'>Lysine access channel</scene> comprised of Tyr335 and Tyr337. Feeding the histone into the active site is initially difficult; however, once in the active site, the alkyl part of the histone chain is stabilized by the <scene name='81/811092/Hydrophobic_packing/4'>hydrophobic binding pocket</scene>, and polar residues are stabilized by hydrogen bonding interactions on the surface. The Tyr335 and Tyr337 are also essential for stabilization of histone chain via hydrogen bonding. The <scene name='81/811092/Active_site_w_water/3'>active site</scene> itself contains the cofactor [https://en.wikipedia.org/wiki/S-Adenosyl_methionine S-adenosyl methionine (SAM)] which donates the methyl group in the reaction. <ref name="Xiao" /> In the active site scene, the structures depict the post-reaction result, where the Lys has been methylated and SAM has been converted to S-adenosyl homocysteine (SAH). |
| | + | |
| | + | [[Image:KMT_Mechanism_jpg.jpeg|200px|left|thumb|Figure 2. KMT Mechanism]] |
| | + | |
| | + | The reaction is catalyzed by Tyr305, Tyr245, carbonyl oxygens of the main chain in residues Ala295 and Ser290. Tyr305 and the carbonyl oxygens of Ala295 and Set 290 coordinate with a water molecule to in turn coordinate with one of the hydrogens off the nitrogen of the lysine, while oxygen of Tyr245 pulls on the other hydrogen of the nitrogen. Both of these actions allow nitrogen to become more nucleophilic and attack the carbon of the methyl group on the SAM, which is attached to a positively charged sulfur. The methyl group is then transferred and charge on the sulfur is resolved; SAM has been converted to SAH. <ref name="Xiao" /> |
| | + | |
| | + | |
| | + | |
| | + | ===The C-Terminal Domain=== |
| | + | |
| | + | The C-terminal domain of lysine methyltransferase is very important for the catalytic activity of the enzyme. The structures of the <scene name='81/811091/C_terminal_domain/1'>C-terminal domain (residues 345-366)</scene> serve the role of stabilizing the structures in the <scene name='81/811091/C_terminal_domain/16'>SET7 domain (residues 193-344)</scene> in the correct orientation for a reaction in the active site.<ref name="Xiao" /> Hydrophobic interactions in the C-terminal domain are mainly responsible for stabilizing the access channel for the lysine methylation site on histone H3. Residues 337-349 create a <scene name='81/811086/Beta-hairpin/1'>beta-hairpin</scene> that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. Furthermore, the hydrophobic packing of alpha-helix 3 against beta-sheet 19, specifically <scene name='81/811091/C_terminal_domain/13'>residues Leu357 and Phe 299</scene>, stabilize the orientation of the <scene name='81/811091/C_terminal_domain/20'>SAM cofactor</scene> so that its methyl donating group is oriented toward the lysine access channel. The orientation of the SAM cofactor is further stabilized with its hydrophobic interactions with C-terminal domain residues <scene name='81/811092/C_terminal_domain/1'>Glu356 and Trp352</scene>. <ref name="Xiao" /> |
| | + | |
| | + | |
| | + | |
| | + | ===The N-terminal Domain=== |
| | + | |
| | + | [[Image:Clustal.png|200px|right|thumb|Figure 1. Clustal alignment of N-terminals]] |
| | + | |
| | + | Though a highly conserved region, the <scene name='81/811092/N_term_domain/1'>N-terminal domain</scene> of SET7 is notably far from the active site and has not been shown to be involved in enzyme activity or participate in substrate binding. <ref name="Kwon">PMID: 12514135</ref> With deletion of the N-terminal domain, studies have shown this modification does not affect SET7 activity. <ref name="Xiao" /> Though not essential for catalytic activity, the N-terminal domain may interact with other small molecules or proteins to act as an allosteric regulator region to the C-terminal domain. <ref name="Kwon" /> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | ==Inhibitors== |
| | + | |
| | + | |
| | + | |
| | + | SET7/9’s structure and function has been studied extensively because of its role in transcription <ref name="Takemoto">PMID:27088648</ref>. In the past few years it has been identified to methylate genes involved in multiple diseases; making it a potential candidate for drug inhibition. <ref name="Tamura">PMID:29723250</ref> Two compounds that have been found to inhibit SET7/9 in certain cells in vitro are Sinefungin and Cyproheptadine. Each inhibitor acts on the catalytic center of SET7/9, however their mechanisms of inhibition and possible medical relevancies differ greatly. |
| | + | |
| | + | |
| | + | |
| | + | ===Sinefungin=== |
| | + | |
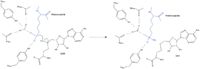
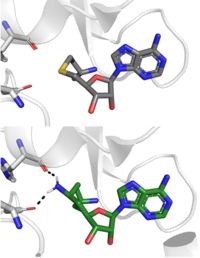
| | + | Sinefungin is a potent methyltransferase inhibitor that is a natural nucleoside isolated from the [https://www.britannica.com/science/Streptomyces "Streptomyces"] species. <ref name="Tamura" /> Also referred to as adenosyl-ornithine, it is the delta (5’ adenosyl) derivative of [https://en.wikipedia.org/wiki/Ornithine ornithine] and a [https://en.wikipedia.org/wiki/Structural_analog structural analog] of S-adenosylmethionine. Sinefugin is unique because it inhibits where the cofactor binds rather than where the substrate binds like a typical competitive inhibitor. Sinefungin is more stable bound in the active site than SAH due to the ability to create two additional hydrogen bonds to its amine group that are not possible with SAH’s sulfur. |
| | + | |
| | + | |
| | + | |
| | + | [[Image: SinSAH.jpg|200 px| right| thumb|SAH (grey) and Sinefungin (green) in the peptide binding pocket. The nitrogen group of sinefungin makes 2 double bonds to the main chain carbonyls of Arg265 and His293. Sinefungin was created using PDB: 1O9S and mutating the sulfur of SAH]] |
| | + | |
| | + | |
| | + | |
| | + | Sinefungin has been used experimentally to inhibit the SET 7/9 protein on [https://www.sciencedirect.com/topics/medicine-and-dentistry/peritoneal-fibrosis peritoneal fibrosis] in mice and in human peritoneal mesothelial cells. <ref name="Tamura" /> SET 7/9 is involved in peritoneal fibrosis because it mono-methylates [https://epigenie.com/key-epigenetic-players/histone-proteins-and-modifications/histone-h3k4/ H3K4], which activates the transcription of fibrosis related genes. The administration of Sinefungin to mice in vitro resulted in decreased levels of methylated H3K4 (H3K4me1) protein, as well as suppressed peritoneal cell density and thickening. The decreased levels of H3K4me1 suggest that the methylation of H3K4 was inhibited by Sinefungin, as well as that inhibiting SET7/9 ameliorates peritoneal fibrosis. |
| | + | |
| | + | |
| | + | |
| | + | ===Cyproheptadine=== |
| | + | |
| | + | Another inhibitor of SET 7/9 is <scene name='81/811086/Cyproheptadine/3'>cyproheptadine</scene>, a clinically-approved anti-allergy drug that was originally developed as a serotonin and histimine. <ref name="Takemoto" /> The cyproheptadine-SET 7/9 complex was crystallized via X-ray diffraction at 2.005 Å with methylated cofactor SAM and with cydroheptadine. Unlike Sinefungin, it is a traditional competitor and competitive with the peptide substrates as it binds to the peptide-binding site. When cyproheptadine binds to the substrate site, the nitrogen of the [https://www.koeichem.com/en/product/index.php/item?cell003=Amines&cell004=Piperidine+derivatives&page=8&name=N-Methylpiperidine&id=116&label=1 methylpiperdine] ring of cyproheptadine forms a hydrogen bond with Thr286 as well as hydrophobic and interactions with the residues surrounding its binding site. The binding of cyproheptadine to the catalytic site causes conformational changes of residue Tyr337, an important residue for the formation of the lysine access channel. This movement subsequently causes a conformational change of the <scene name='81/811086/Betahairincypr/1'>beta hairpin</scene>. The <scene name='81/811086/Betahairpincyp/3'>residues 337-349</scene> conformational change ultimately generates a large hole adjacent to the lysine access channel, as well as the shift of the C-terminal helix.<ref name="Takemoto" /> |
| | + | |
| | + | With the revelation of its inhibitory effects on SET7/9, cyproheptadine was used in vitro to treat breast cancer cells ([https://en.wikipedia.org/wiki/MCF-7 MCF-7] cells). SET 7/9's non-histone activities include the methylation of the estrogen receptor α (ERα), a nuclear receptor and a transcription factor responsible for estrogen-responsive gene regulation. The expression and transcriptional activity of ERα is involved in the carcinogenesis of 70% of breast cancers, making it a major target for hormone therapy. Researchers found that treating the MCF7 cells with cyproheptadine decreased ERα's expression and transcriptional activity which therefore inhibited the estrogen-dependent cell growth. These findings suggest that cyproheptadine could possibly be repurposed to breast cancer therapy in the future.<ref name="Takemoto" /> |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | </StructureSection> |
| | + | |
| | + | |
| | + | |
| | + | == References == |
| | + | |
| | + | <references/> |
| | + | |
| | + | |
| | + | |
| | + | ==Student Contributors== |
| | + | |
| | + | Lauryn Padgett, |
| | + | |
| | + | Alexandra Pentala, |
| | + | |
| | + | Madeleine Wilson |