We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Kyle Burton/Sandbox1
From Proteopedia
< User:Kyle Burton(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 7: | Line 7: | ||
== Structure == | == Structure == | ||
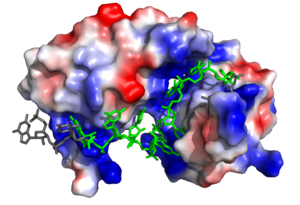
[[Image:Sex lethal protein electrostatic surface representation.png|300px|left|thumb| '''Figure 2.''' Sxl showing the electropositive binding pocket and the bound RNA ligand. Pre-mRNA residues binding to Sxl shown in green, non-binding residues shown in grey. Structure shown is [https://www.rcsb.org/structure/1b7f PDB:1b7f]. Figure created in PyMol.]] | [[Image:Sex lethal protein electrostatic surface representation.png|300px|left|thumb| '''Figure 2.''' Sxl showing the electropositive binding pocket and the bound RNA ligand. Pre-mRNA residues binding to Sxl shown in green, non-binding residues shown in grey. Structure shown is [https://www.rcsb.org/structure/1b7f PDB:1b7f]. Figure created in PyMol.]] | ||
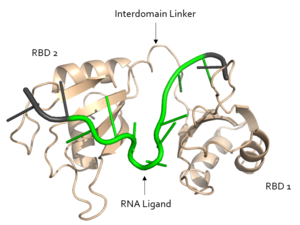
| - | Sxl is composed of two asymmetric RNA binding domains (RBD1 and RBD2) which recognize a poly-uridine site in the pre-mRNA transcript<ref name="Handa"/>. <scene name='78/783145/Secondary_structure/3'>Both RBDs</scene> are comprised of two [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] and one antiparallel four-stranded [https://en.wikipedia.org/wiki/Beta_sheet β sheet]<ref name="Handa"/> containing the [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motif](Fig. 1). The β sheets face each other, lining the V-shaped cleft<ref name="Handa"/>, shown in sand in Fig. 1. The inter-domain linker, shown in sand in Fig. 1, forms a distorted 3<sub>10</sub> helix which helps form the V-shaped cleft into which the pre-mRNA sequence binds<ref name="Handa"/><ref name="Black">doi: 10.1146/annurev.biochem.72.121801.161720</ref>. Sxl binds to UGUUUUUUU sequence of GUUGUUUUUUUU in the ''tra'' pre-mRNA<ref name="Handa"/><ref name="Black"/>. RBD1 binds U6-U11 and RBD2 binds U3, G4, and U5. Figure 1 shows bound pre-mRNA residues in green and non-bound pre-mRNA residues in grey. Although the two RBDs do not interact with each other, this nine-ribonucleotide sequence must be recognized continuously to allow Sxl to bind, preventing U2AF from binding at the 3’ splice site<ref name="Handa"/>. The binding of Sxl to the pre-mRNA occurs in an electropositive pocket (shown in blue in Fig. 2) due to extensive interactions with the RNA phosphate backbone and negatively charged residues<ref name="Handa"/>. | + | Sxl is composed of two asymmetric RNA binding domains (RBD1 and RBD2) which recognize a poly-uridine site in the pre-mRNA transcript<ref name="Handa"/>. <scene name='78/783145/Secondary_structure/3'>Both RBDs</scene> are comprised of two [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] and one antiparallel four-stranded [https://en.wikipedia.org/wiki/Beta_sheet β sheet]<ref name="Handa"/> containing the [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motif](Fig. 1). The β sheets face each other, lining the V-shaped cleft<ref name="Handa"/>, shown in sand in Fig. 1. The inter-domain linker, shown in sand in Fig. 1, forms a distorted 3<sub>10</sub> helix which helps form the V-shaped cleft into which the pre-mRNA sequence binds<ref name="Handa"/><ref name="Black">doi: 10.1146/annurev.biochem.72.121801.161720</ref>. Sxl binds to UGUUUUUUU sequence of GUUGUUUUUUUU in the ''tra'' pre-mRNA<ref name="Handa"/><ref name="Black"/>. RBD1 binds U6-U11 and RBD2 binds U3, G4, and U5. Figure 1 shows bound pre-mRNA residues in green and non-bound pre-mRNA residues in grey. Although the two RBDs do not interact with each other, this nine-ribonucleotide sequence must be recognized continuously to allow Sxl to bind, preventing U2AF from binding at the 3’ splice site<ref name="Handa"/>. The binding of Sxl to the pre-mRNA occurs in an electropositive pocket (shown in blue in Fig. 2) due to extensive interactions with the RNA phosphate backbone and negatively charged residues<ref name="Handa"/>. Because non-RBD residues are not essential to Sxl's function, there is some variation in the ''sxl'' gene of other drosopholids, though the RBD residues are highly conserved among all drosopholids<ref name="Penalva"/>. |
=== Structural Basis for Recognition of Poly-U Sequences === | === Structural Basis for Recognition of Poly-U Sequences === | ||
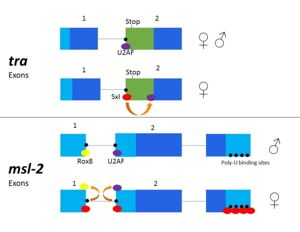
| - | The structural interactions with regards to the targeting of the 5' splice site and of its own mRNA transcript are much less understood than the competition of <scene name='78/783145/Sxl/ | + | The structural interactions with regards to the targeting of the 5' splice site and of its own mRNA transcript are much less understood than the competition of <scene name='78/783145/Sxl/2'>Sxl</scene> with U2AF at the 3' splice site. All the RNA-protein interactions described here refer to ''tra''-Sxl interactions<ref name="Handa"/>. There are no published crystal structures of the Sxl-''msl-2'' complex, but Sxl recognizes the same poly-U site in both ''tra'' and ''msl-2''. |
The <scene name='78/783145/Arg_252_interaction_with_u3_g4/6'>R252 interaction with U3 and G4</scene> is crucial to pre-mRNA binding; a mutation of R252 to alanine eliminated the ability of Sxl to bind RNA<ref name="original">PMID: 9398148</ref>. | The <scene name='78/783145/Arg_252_interaction_with_u3_g4/6'>R252 interaction with U3 and G4</scene> is crucial to pre-mRNA binding; a mutation of R252 to alanine eliminated the ability of Sxl to bind RNA<ref name="original">PMID: 9398148</ref>. | ||
| Line 18: | Line 18: | ||
The U6 residue is recognized as part of the RNA <scene name='78/783145/U5_u6_u7_loop/13'>loop at U5, U6, and U7</scene> by R195. The R195 amide hydrogen-bonds to the O2' of U6 and the U6 N3H hydrogen bonds to the R195 carbonyl oxygen. | The U6 residue is recognized as part of the RNA <scene name='78/783145/U5_u6_u7_loop/13'>loop at U5, U6, and U7</scene> by R195. The R195 amide hydrogen-bonds to the O2' of U6 and the U6 N3H hydrogen bonds to the R195 carbonyl oxygen. | ||
| - | In the <scene name='78/783145/U5_u6_u7_loop/ | + | In the <scene name='78/783145/U5_u6_u7_loop/14'>RNA loop</scene>, the U7 and U8 bases are involved in <scene name='78/783145/U7_u8_stacking/3'>π stacking</scene>, stabilizing the 3' endo conformation of the U8 sugar. U8 is further stabilized via hydrogen bonding <scene name='78/783145/U8_with_s165_and_y166/5'>interactions with S165 and Y166</scene>, where the nitrogens of the uridine ring hydrogen bond to the respective carbonyl oxygens of S165 and Y166. |
| - | <scene name='78/783145/N130_interaction_with_u9/ | + | <scene name='78/783145/N130_interaction_with_u9/5'>U9</scene> is recognized by the interdomain linker. This interaction is permissible due to free rotation in the N130 side chain, allowing hydrogen bonding between the N130 side chain and a phosphate oxygen of U9. U9 is further stabilized by a second <scene name='78/783145/U9_with_interdomain_linker/1'>an ion-dipole interaction</scene> between the U9 O2' and the side chain of R202 and the U9 O4' and the K197 side chain. |
U9 facilitates the stabilization of U10, which is also recognized by the interdomain linker. A phosphate oxygen of <scene name='78/783145/Arg_258_interaction_w_u9_u10/3'>R258 interacts with U9 and U10</scene> to form a [https://en.wikipedia.org/wiki/Salt_bridge salt bridge]. | U9 facilitates the stabilization of U10, which is also recognized by the interdomain linker. A phosphate oxygen of <scene name='78/783145/Arg_258_interaction_w_u9_u10/3'>R258 interacts with U9 and U10</scene> to form a [https://en.wikipedia.org/wiki/Salt_bridge salt bridge]. | ||
Current revision
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999 Apr 15;398(6728):579-85. PMID:10217141 doi:10.1038/19242
- ↑ 2.0 2.1 2.2 2.3 2.4 Penalva L, Sanchez L. RNA Binding Protein Sex-Lethal (Sxl) and Control of Drosophila Sex Determination and Dosage Compensation. Microbiol Mol Biol Rev.;67(3):343-356. doi: 10.1128/MMBR.67.3.343–359.2003
- ↑ 3.0 3.1 3.2 Bashaw GJ, Baker BS. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development. 1995 Oct;121(10):3245-58. PMID:7588059
- ↑ 4.0 4.1 4.2 4.3 Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995 Jun 16;81(6):867-77. PMID:7781064
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336. doi: 10.1146/annurev.biochem.72.121801.161720., Epub 2003 Feb 27. PMID:12626338 doi:http://dx.doi.org/10.1146/annurev.biochem.72.121801.161720
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Georgiev P, Chlamydas S, Akhtar A. Drosophila dosage compensation: males are from Mars, females are from Venus. Fly (Austin). 2011 Apr-Jun;5(2):147-54. Epub 2011 Apr 1. PMID:21339706
- ↑ Lee AL, Volkman BF, Robertson SA, Rudner DZ, Barbash DA, Cline TW, Kanaar R, Rio DC, Wemmer DE. Chemical shift mapping of the RNA-binding interface of the multiple-RBD protein sex-lethal. Biochemistry. 1997 Nov 25;36(47):14306-17. doi: 10.1021/bi970830y. PMID:9398148 doi:http://dx.doi.org/10.1021/bi970830y
- ↑ 8.0 8.1 Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991 Apr 19;65(2):229-39. PMID:2015624
- ↑ Gebauer F, Merendino L, Hentze MW, Valcarcel J. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA. 1998 Feb;4(2):142-50. PMID:9570314
- ↑ Inoue M, Muto Y, Sakamoto H, Kigawa T, Takio K, Shimura Y, Yokoyama S. A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal. J Mol Biol. 1997 Sep 12;272(1):82-94. PMID:9299339 doi:10.1006/jmbi.1997.1213