Nuclear polyadenylated RNA-binding protein
From Proteopedia
(Difference between revisions)
| (14 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='2cjk' size='400' side='right' caption=' | + | <StructureSection load='2cjk' size='400' side='right' caption='Yeast nuclear polyadenylated RNA-binding protein 4 (Hrp1) complex with RNA (PDB code [[2cjk]])' scene='78/781945/Hrp1_and_pee/1'> |
=Introduction= | =Introduction= | ||
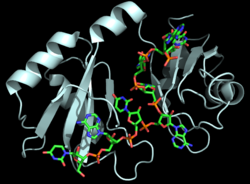
[[Image:Hrp1 fig1 cropped.png|250 px|right|thumb|Figure 1: Cartoon representation of the Hrp1-PEE complex. The RNA is shown as a stick model and is colored by element.]] | [[Image:Hrp1 fig1 cropped.png|250 px|right|thumb|Figure 1: Cartoon representation of the Hrp1-PEE complex. The RNA is shown as a stick model and is colored by element.]] | ||
| - | Hrp1 is a [https://en.wikipedia.org/wiki/Polyadenylation polyadenylation] factor found in ''Saccharomyces cerevisiae'' (yeast) <ref name="GM3H">PMID: 16794580</ref>. Hrp1 specifically recognizes and binds to an RNA sequence in the [https://en.wikipedia.org/wiki/Three_prime_untranslated_region 3'UTR] of the [https://en.wikipedia.org/wiki/Messenger_RNA messenger RNA (mRNA)] upstream from the cleavage site called the polyadenylation enhancement element (PEE) (Figure 1) <ref name="GM3H"/>. Upon binding to the RNA, Hrp1 helps recruit additional proteins necessary for the cleavage and polyadenylation of the RNA molecule <ref name="GM3H"/>. Although Hrp1 shares several common features with other RNA-binding proteins, the unique structural features of the Hrp1-PEE complex reveals the mechanism by which Hrp1 is able to recognize and bind to its specific RNA sequence at the atomic level <ref name="GM3H"/>. Hrp1 was discovered when Cleavage Factor I (CF I) was purified and separated into its two components, CF IA and CF IB. CF IB is a single 73 kDa polypeptide. The polypeptide was digested and two tryptic peptides were obtained for sequencing. The sequences were aligned via a database, and Hrp1 was determined to be a perfect match. Hrp1 of CF IB interacts with Rna14 and Rna15 of CF IA<ref name="RNA15"/> to form a protein complex that aids in cleavage and polyadenylation of pre-mRNA and transport of mature mRNA from the nucleus<ref name="KHSZ">PMID: 9334319</ref>. | + | '''Nuclear polyadenylated RNA-binding proteins''' (NAB) are yeast proteins which may be required for packaging pre-mRNAs into ribonucleoprotein structures amenable to efficient nuclear RNA processing<ref>PMID:7962083</ref>.<br /> |
| + | *'''Nab1''' is involved in the nucleocytoplasmic protein shuttling and pre_RNA processing<ref>PMID:7962083</ref>. <br /> | ||
| + | *'''Nab2''' is essential for cell viability<ref>PMID:8474438</ref>. <br /> | ||
| + | *'''Nab3''' interacts with the nascent RNA transcript and [[RNA polymerase]] II<ref>PMID:23192344</ref>. <br /> | ||
| + | *'''Hrp1''' or '''Nab4''' is a [https://en.wikipedia.org/wiki/Polyadenylation polyadenylation] factor found in ''Saccharomyces cerevisiae'' (yeast) <ref name="GM3H">PMID: 16794580</ref>. Hrp1 specifically recognizes and binds to an RNA sequence in the [https://en.wikipedia.org/wiki/Three_prime_untranslated_region 3'UTR] of the [https://en.wikipedia.org/wiki/Messenger_RNA messenger RNA (mRNA)] upstream from the cleavage site called the polyadenylation enhancement element (PEE) (Figure 1) <ref name="GM3H"/>. Upon binding to the RNA, Hrp1 helps recruit additional proteins necessary for the cleavage and polyadenylation of the RNA molecule <ref name="GM3H"/>. Although Hrp1 shares several common features with other RNA-binding proteins, the unique structural features of the Hrp1-PEE complex reveals the mechanism by which Hrp1 is able to recognize and bind to its specific RNA sequence at the atomic level <ref name="GM3H"/>. Hrp1 was discovered when Cleavage Factor I (CF I) was purified and separated into its two components, CF IA and CF IB. CF IB is a single 73 kDa polypeptide. The polypeptide was digested and two tryptic peptides were obtained for sequencing. The sequences were aligned via a database, and Hrp1 was determined to be a perfect match. Hrp1 of CF IB interacts with Rna14 and Rna15 of CF IA<ref name="RNA15"/> to form a protein complex that aids in cleavage and polyadenylation of pre-mRNA and transport of mature mRNA from the nucleus<ref name="KHSZ">PMID: 9334319</ref>. | ||
=Structure= | =Structure= | ||
==General Features== | ==General Features== | ||
| - | Hrp1 is a single-stranded [https://en.wikipedia.org/wiki/RNA-binding_protein RNA-binding protein] composed of two RNP-type [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA-binding domains (RBDs)] arranged in tandem with a typical ßαßßαß architecture <ref name="GM3H"/>. The two RBDs have similar topolgies, both containing a central [https://en.wikipedia.org/wiki/Beta_sheet antiparallel] four-stranded <scene name='78/783765/Beta_sheet/1'>ß-sheet</scene> with two [https://en.wikipedia.org/wiki/Alpha_helix α-helices] running across one face <ref name="GM3H"/>. The two RBDs associate to form a deep and positively charged <scene name='78/781960/Hrp1-rna_interface_surface/ | + | '''Hrp1''' is a single-stranded [https://en.wikipedia.org/wiki/RNA-binding_protein RNA-binding protein] composed of two RNP-type [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA-binding domains (RBDs)] arranged in tandem with a typical ßαßßαß architecture <ref name="GM3H"/>. The two RBDs have similar topolgies, both containing a central [https://en.wikipedia.org/wiki/Beta_sheet antiparallel] four-stranded <scene name='78/783765/Beta_sheet/1'>ß-sheet</scene> with two [https://en.wikipedia.org/wiki/Alpha_helix α-helices] running across one face <ref name="GM3H"/>. The β-strands of each βαβ domain are linked via hydrogen bonding between conserved residues, <scene name='78/783765/L166_g201/4'>Leu166 and Gly201</scene>. The two RBDs associate to form a deep and positively charged <scene name='78/781960/Hrp1-rna_interface_surface/3'>cleft</scene>, which constitutes the binding site for the RNA molecule <ref name="GM3H"/>. |
==Hrp1-RNA Interactions== | ==Hrp1-RNA Interactions== | ||
| - | The interface between Hrp1 and its target RNA sequence is dominated by interactions between key aromatic residues and RNA nucleobases <ref name="GM3H"/>. Only six RNA bases, an <scene name='78/781952/Ua_repeats/ | + | The interface between Hrp1 and its target RNA sequence is dominated by interactions between key aromatic residues and RNA nucleobases <ref name="GM3H"/>. Only six RNA bases, an <scene name='78/781952/Ua_repeats/2'>(AU)3</scene> repeat, act as the PEE and form specific contacts with Hrp1 <ref name="GM3H"/>. The kinked conformation around Ade4 is uncommon for RNA alone, and may be adopted by the RNA for specific interactions with Hrp1. Ade4 is part of a crucial interaction with Trp168 which will be discussed later, and could explain the adoption of the kinked conformation. Hydrophilic residues of Hrp1 provide base specificity through hydrogen bonding <ref name="GM3H"/>. Most of the key residues that interact with the RNA can be found in the ß-sheet region of Hrp1; however, loops and the interdomain linker are also essential for Hrp1-RNA recognition <ref name="GM3H"/>. Perhaps the most important Hrp1-RNA interaction is the <scene name='78/783765/Ade4-trp168/4'>interaction between Ade4 and Trp168</scene> <ref name="GM3H"/>. In this case, Trp168 stacks on Ade4 and forms crucial base-specific hydrogen bonds such as the H-bond between N7 of Ade4 and amide hydrogren of Trp168 <ref name="GM3H"/>. This interaction is a unique feature of the Hrp1-PEE complex and has not been found in any other single-stranded RNA-binding proteins with two canonical RBDs <ref name="GM3H"/>. It is also worth noting that a second Hrp1 residue is critical to holding Ade4 in place, <scene name='78/781945/Lys226-ade4-trp168/3'>Lys226</scene>, which stabilizes Ade4 likely via a [https://en.wikipedia.org/wiki/Cation%E2%80%93pi_interaction cation–π interaction]. As displayed in the structure, the lysine cationic nitrogen is approximately 4Å away from the pi system, which is within the 6Å range associated with cation-pi interactions. A third contributor, <scene name='78/783765/Phe204_and_u7_interaction/2'>Phe204</scene>, also stacks with Ura7 to aid in RNA recognition and binding <ref name="GM3H"/>. |
==RBD-RBD Interactions and the Linker Region== | ==RBD-RBD Interactions and the Linker Region== | ||
| - | As mentioned above, Hrp1 is composed of two RBDs. The RBDs are connected by a <scene name='78/783765/Linker/3'>linker region</scene> (a short two-turn α-helix), which also contains a crucial residue for RNA binding. Ile234 in the linker region holds Ade6 in place in order to ensure proper <scene name='78/781945/Linker_rna/2'>via van der Waals contacts</scene> with the nearby Phe162. Experimental evidence from protein [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance nuclear magnetic resonance (NMR)] data <ref name="GM3H"/> suggests that the two RBDs move independently prior to binding the PEE. Upon binding the PEE, the linker region adopts a short helical structure to rigidly hold the RBDs in place relative to each other. Aside from the linker helix, the only other interaction between the RBDs is <scene name='78/781945/Interaction_between_domains/8'>a single salt bridge</scene> between Lys231 and Asp271 <ref name="GM3H"/>. | + | As mentioned above, '''Hrp1''' is composed of two RBDs. The RBDs are connected by a <scene name='78/783765/Linker/3'>linker region</scene> (a short two-turn α-helix), which also contains a crucial residue for RNA binding. Ile234 in the linker region holds Ade6 in place in order to ensure proper <scene name='78/781945/Linker_rna/2'>via van der Waals contacts</scene> with the nearby Phe162. Experimental evidence from protein [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance nuclear magnetic resonance (NMR)] data <ref name="GM3H"/> suggests that the two RBDs move independently prior to binding the PEE. Upon binding the PEE, the linker region adopts a short helical structure to rigidly hold the RBDs in place relative to each other. Aside from the linker helix, the only other interaction between the RBDs is <scene name='78/781945/Interaction_between_domains/8'>a single salt bridge</scene> between Lys231 and Asp271 <ref name="GM3H"/>. |
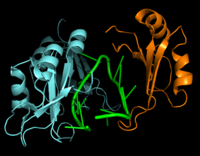
[[Image:Hrp1 RNA15 Cropped.png|200 px|left|thumb|Figure 2: Interaction between Hrp1 (blue), RNA15 (orange) and RNA (green).]] | [[Image:Hrp1 RNA15 Cropped.png|200 px|left|thumb|Figure 2: Interaction between Hrp1 (blue), RNA15 (orange) and RNA (green).]] | ||
| Line 22: | Line 26: | ||
=Relationship to other proteins= | =Relationship to other proteins= | ||
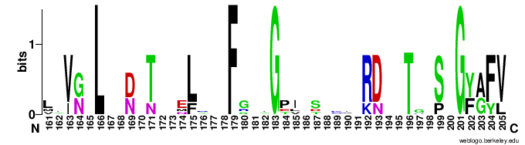
| - | The RNP-type RBD is found in many proteins involved in post-transcriptional [https://en.wikipedia.org/wiki/Post-transcriptional_modification pre-mRNA processing] (5'-end capping, splicing, 3'-end cleavage and polyadenylation, and transport from the nucleus)<ref name="RRMB">PMID: 18515081</ref>. The unique RBD of Hrp1 enables the protein to bind an RNA sequence that differs in both length and content from the RNA sequences of other RNA-binding and mRNA processing proteins such as [http://proteopedia.org/wiki/index.php/2sxl sex lethal], [https://en.wikipedia.org/wiki/Poly(A)-binding_protein Poly (A)-binding protein (PABP)], and [http://proteopedia.org/wiki/index.php/1fxl HuD] <ref name="GM3H"/>. These proteins, in addition to Hrp1 and | + | The RNP-type RBD is found in many proteins involved in post-transcriptional [https://en.wikipedia.org/wiki/Post-transcriptional_modification pre-mRNA processing] (5'-end capping, splicing, 3'-end cleavage and polyadenylation, and transport from the nucleus)<ref name="RRMB">PMID: 18515081</ref>. The unique RBD of Hrp1 enables the protein to bind an RNA sequence that differs in both length and content from the RNA sequences of other RNA-binding and mRNA processing proteins such as [http://proteopedia.org/wiki/index.php/2sxl sex lethal], [https://en.wikipedia.org/wiki/Poly(A)-binding_protein Poly (A)-binding protein (PABP)], and [http://proteopedia.org/wiki/index.php/1fxl HuD] <ref name="GM3H"/>. These proteins, in addition to Hrp1 and [http://proteopedia.org/wiki/index.php/2i2y SRp20], contain conserved hydrophobic residues which contribute to hydrophobic interactions between secondary structures of the proteins. Each protein also contains conserved residues L166 and G201 which form a hydrogen bond, linking the β-sheets in the βαβ complex of Hrp1 (Figure 3). [[Image:Conserved_Hrp1_sequence_logo.png|525 px|center|thumb|Figure 3: Sequence logo for residues 161-205 of Hrp1.]] Like Hrp1, each of these proteins belongs to the class of single-stranded proteins composed of two canonical RBDs; however, each protein is differentiated by respective target RNA sequences, interactions with RNA at the atomic level, and interdomain contacts <ref name="GM3H"/>. Hrp1 is unique in that HuD, sex lethal, and PABP all contain at least one intra-RNA base-base stacking interaction, a feature that is lacking in the Hrp1-PEE complex <ref name="GM3H"/>. It is possible that the intra-RNA interactions found in these other protein-RNA complexes is replaced by the crucial Trp168-Ade4 stacking interaction found in the Hrp1 complex <ref name="GM3H"/>. This may help explain why the Hrp1-RNA interface involves only 6 nucleotides whereas PABP, sex lethal, and HuD require a longer 8-10 nucleotide sequence in the RNA binding pocket <ref name="GM3H"/>. |
</StructureSection> | </StructureSection> | ||
| + | == 3D Structures of nuclear polyadenylated RNA-binding protein == | ||
| + | |||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *Nuclear polyadenylated RNA-binding protein 1 or nucleolar protein 3 | ||
| + | |||
| + | **[[2jvo]], [[2osq]], [[7qdd]] – yNAB1 RRM-1 domain 116-201 – yeast - NMR<br /> | ||
| + | **[[2osr]], [[2jvr]] – yNAB1 RRM-2 domain 194-280 - NMR<br /> | ||
| + | **[[7qde]], [[8b8s]] – yNAB1 RRM1-RRM2 domain + RNA - NMR<br /> | ||
| + | |||
| + | *Nuclear polyadenylated RNA-binding protein 2 | ||
| + | |||
| + | **[[2v75]] – yNab2 N-terminal 1-106<br /> | ||
| + | **[[2v75]], [[2jps]] – yNab2 N-terminal - NMR<br /> | ||
| + | **[[3lcn]] – yNab2 N-terminal + Gfd1 <br /> | ||
| + | **[[3zj1]] – yNab2 zinc finger 1+2 253-333 - NMR<br /> | ||
| + | **[[3zj2]] – yNab2 zinc finger 3+4 333-401 - NMR<br /> | ||
| + | **[[2lhn]] – yNab2 zinc finger 5-7 409-483 - NMR<br /> | ||
| + | **[[5l2l]] – yNab2 zinc finger 5-7 407-483 + RNA<br /> | ||
| + | **[[4jlq]] – yNab2 205-242 + transportin 1<br /> | ||
| + | |||
| + | *Nuclear polyadenylated RNA-binding protein 3 | ||
| + | |||
| + | **[[2xnq]] – yNab3 RNA recognition 329-404 <br /> | ||
| + | **[[2kvi]] – yNab3 RNA recognition - NMR<br /> | ||
| + | **[[2xnr]] – yNab3 RNA recognition + RNA<br /> | ||
| + | **[[2l41]] – yNab3 RNA recognition + RNA - NMR<br /> | ||
| + | |||
| + | *Nuclear polyadenylated RNA-binding protein 4 | ||
| + | **[[2cjk]] – yNab4 RNA recognition 156-322 + RNA - NMR<br /> | ||
| + | **[[2km8]] – yNab4 RNA recognition 156-322 + mRNA 3’-end-processing protein + RNA - NMR<br /> | ||
| + | }} | ||
=References= | =References= | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
3D Structures of nuclear polyadenylated RNA-binding protein
Updated on 20-July-2023
References
- ↑ Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson MS. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994 Dec;127(5):1173-84. PMID:7962083
- ↑ Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson MS. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994 Dec;127(5):1173-84. PMID:7962083
- ↑ Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993 May;13(5):2730-41. PMID:8474438
- ↑ Loya TJ, O'Rourke TW, Reines D. Yeast Nab3 protein contains a self-assembly domain found in human heterogeneous nuclear ribonucleoprotein-C (hnRNP-C) that is necessary for transcription termination. J Biol Chem. 2013 Jan 25;288(4):2111-7. doi: 10.1074/jbc.M112.430678. Epub 2012, Nov 28. PMID:23192344 doi:http://dx.doi.org/10.1074/jbc.M112.430678
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 5.17 5.18 5.19 5.20 5.21 Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3'UTR recognition by Hrp1. EMBO J. 2006 Jul 12;25(13):3167-78. Epub 2006 Jun 22. PMID:16794580
- ↑ 6.0 6.1 6.2 6.3 6.4 Leeper TC, Qu X, Lu C, Moore C, Varani G. Novel protein-protein contacts facilitate mRNA 3'-processing signal recognition by Rna15 and Hrp1. J Mol Biol. 2010 Aug 20;401(3):334-49. Epub 2010 Jun 19. PMID:20600122 doi:10.1016/j.jmb.2010.06.032
- ↑ Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3'-end formation in yeast. Genes Dev. 1997 Oct 1;11(19):2545-56. PMID:9334319
- ↑ Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008 Jun;18(3):290-8. doi: 10.1016/j.sbi.2008.04.002. PMID:18515081 doi:http://dx.doi.org/10.1016/j.sbi.2008.04.002
Proteopedia Page Contributors and Editors (what is this?)
Cory A. Wuerch, Matthew Douglas Moore, Savannah Davis, Michal Harel, Jaime Prilusky