Introduction
is a homodimer (150kDa in bacteria and 170kDa in human) and is classified as a class II
Aminoacyl-tRNA synthetase enzymes. These ancient enzymes primary function are to add the respective amino acid to the respective transfer Ribonucleic Acid (tRNA-AA), a necessity prep for the protein synthesis pathway
[1]. As the name implies, TARS function is to add Threonine amino acid (Thr) to threonine specific tRNA (tRNA-thr) in the presence of Adenosine triphosphate (ATP) and diavalent metal cation. Below displays the overview of TARS Aminoacylation rxn
[2].
Overall TARS protein rxn. Substrates includes ATP, Thr and tRNA-thr. Although there are multiple forms of TARS in different cells, this protopedia page mainly focuses on the structure of Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) cytoplasmic TARS as their is more crystal structures that identify their domains within the protein.
Mechanism
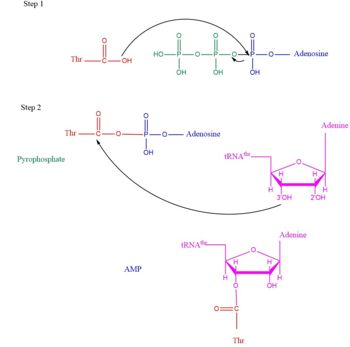
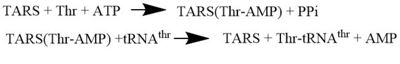
Arrow pushing of Aminoacylation rxn.[3] TARS adds amino acid to tRNA by a two-step mechanism. First the enzyme binds to both and in the catalytic domain to perform an adenylation reaction in which threonyl adenylate (Thr-AMP) is formed and pyrophosphate (PPi) is released as a byproduct. The image to the right displays the binding of adenylate product to the TARS enzyme (PDB entry 1nyq). This is then follow up by a transferring Thr from Adenosine monophosphate (AMP) molecule to 3'OH site of tRNA-thr. [4] The image to the above demonstrates the arrow pushing occurring to generate threonine bound tRNA-thr.
Structural highlights
As mentioned earlier, TARS is homodimer protein found in two forms of a cell, mitochondrial and cytoplasmic. Most of the structures seen will primarly be bacterial cytoplasmic. The protein is mainly classified as a alpha beta protein in both bacteria and eukaryotic cells[5]. Each chain is composed of 4 domain classified by SCOP: [6].
N-terminus (Residue 1-241)
The and is the found in the N-terminus of the TARS enzyme chain. Specific N1 is found from 1-62 residue while N2 domain is found from 63-241 residue(PDB: 1tje). Although located in neighboring regions, the N1-domain and N2-domain deviate dramatically from one another. Structurally, N1-domain has a TGS-fold (short for TARS, GTPase, Spo1 domain as their the most common proteins that present this fold)[7] which has 3 antiparallel beta sheets interacting with an alpha helix causing a formation of roll-like structure (similar to the description by Cath: Ubiquitin-like roll)[8]. The structure has no known function but it is pronounced structure found common within TARS.
This is the opposite of the N2-domain, or sometimes called TARS additional domain. This is ironic as this addition is very important for proofreading TARS activity[9]. TARS N2-domain is described as 2-layer alpha-beta sandwich comprised of mostly alpha helices. Interesting enough, the superfamily of this motif is found to be TARS and Alanyl-tRNA synthetase (AARS)common domain. This domain function similar by discriminating Threonine and Alanine from Serine amino acid (Ser) as serine is similar structure to both amino acids. The editing domain functions first by moving Ser bound tRNA-Thr from the catalytic domain to the by breaking the bonds nucleotide A73, C74, and C75 from the catalytic site allowing the acceptor arm of tRNA-Thr to flip to the editing domain of S12-S13 site. Research on editing hydrolysis propose that residue Tyr 103 plays an important role of guiding acceptor arm towards the editing domain. The Ser bound to tRNA is hydrolyze by a water molecule, interacting with His73, acting as a nucleophile to the alpha carbon of serine, follow by protonation by a 2nd water molecule interacting with carbonyl of Met181 and amide side chain Lys156. The specificity of serine binding to editing site versus threonine is due to steric hindrance by the presents of methyl group in the side chain. What is amazing about the fidility mechanism of AA-tRNA binding is the domain is much similar to (AARS) found in the C-terminus as they share 40% similarity in residues and both select for serine, although AARS also . The subtle difference of the isoforms mitochondrial TARS from cytoplasmic TARS appears in editing as the mitochondrial TARS doesn't have N2-domain and requires interaction to hydrolyze serine [10].
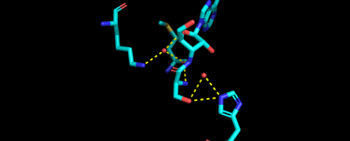
Representative image of editing domain that demonstrates amino acid important in binding and hydrolyzing serine-tRNA(thr). Note, the amino acids highlighted in light blue are the amino acid important for hydrolysis (His-73, Met181, and Lys 156). Also not shown is Tyr103 who plays an important role in changing tRNA acceptor stem from catalytic domain to editing domain. Although separated, both the N2-domain and catalytic domain play an
Catalytic Domain (Residue 243-532)
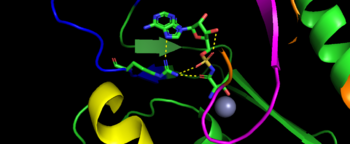
As the name implies, the is the main area of aminoacylation activity. This domain is linked with N2-Domain of TARS by a alpha helix, or a linker helix (residue 225-242)[11]. The domain is architecturally described as a 2-layer alpha beta sandwhich. The importance of the domain is that it has 3 sites of binding: motif 1: ordering loop; motif 2 loop, Motif 3: threonine loop, and motif 4: ATP loop. The main important residues and cofactors within in the catalytic site include: Tyr468, Arg365, His309, Zinc and Magnesium.
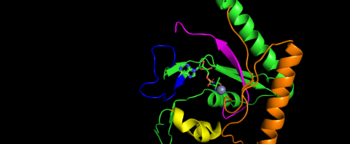
Image represents the motif of S. Aureus TARS. Motif 1 is represent in yellow, motif 2 is represented in blue, motif 3 is represented in orange, and motif 4 is magenta The binding pocket for threonine consistent of Arg363 found in motif 2, Tyr462 found in threonine loop, and zinc divalent ion (Zn2+) coordinating by his-his-cys residues and a water molecule (for S. aureus it was Cys336, His387, and His571; versus E. coli. was Cys334, His385, and His511, thus preventing similar structure molecules that lack hydroxyl group from binding such as valine. The presence of Thr amino acid causes a movement of Tyr468 to move closer to the ordering loop, while the Arg363 is displace to have more interactions with threonine, as well as bind to the alpha phosphate of ATP. Asides Arg363, Glu365 and Arg 375 bind to the gamma and beta phosphates of ATP to stabilize the pyrophosphate product, however both of these residues are not as well conserved as Arg363. The adenine ring is also stabilize in the presence of motif 3 argnine and Phe379. In the presence of Mg2+ (to stabilize the product form), the adenylation reaction occurs, which breaks the bounds seen in Glu365 and Arg 375 to phosphates and the whole ATP loop moving.
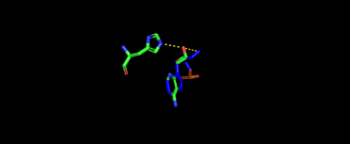
Representative image of catalytic domain binding of threonyl-adenylate. The image includes residue Arg365 binding to threonyl-adenylate representing ATP and Thr amino acylation activity. The 2nd step, formation of thr-tRNA-thr, occurs through tRNA binding to anticodon domain (which is discussed later on) shifting threonine loop and ATP loops, an thus bound threonyl-adenylate, towards the tRNA. The terminal 3'adenine is then hydrogen bound to Tyr462 to stabilize the molecule, follow by subjected to acid-base reaction at the 2'OH site with His309 to catalyze aminoacyl-transfer reaction[12].
Representative image of catalytic domain binding of tRNA. The image includes residue His309 binding to tRNA 3'adenine representing acid-base reaction with 2'OH. Anti-Codon Domain(Residue 533-645)
Lastly, there is the C-terminus domain of the TARS protein, otherwise known as the . This alpha-beta region is conserved through most aaRs class 2 molecules but have a specificity towards binding to tRNA-thr. This domain has a Rossmann-fold, which is 3-layer sandwhich fold necessary for binding of nucleotides, in this case tRNA nucleotides. This domain recognize and bind to tRNA-thr major groove containing BGU anticodon consensus sequence (B: Guanine, Cytosine, or Uracil). Although tRNA binding is important to hold on for aminoacylation, this should not be confused with having any aminoacylation activity.

Representative image of catalytic domain binding of threonyl-adenylate. The image includes residue Arg365 binding to threonyl-adenylate representing ATP and Thr amino acylation activity.
Evolution conservation and related proteins
TARS protein is highly dynamic protein that is necessary for protein synthesis, so it is no surprise to the for the protein. Since both mitochondrial and cytoplasmic activity in eukaryotic, bacterial and archaeal all perform similar function, it makes sense there is no deviation from structure from the catalytic site. The mostly highly conserved amino acid includes R363 (R448 in eukaryotes) which as mentioned earlier is important for ATP binding and is important for the step 1 mechanism of forming threonyl adenylate product[13][14]
Asides catalytic site, the Anticodon binding domain is also highly conserved in most TARS enzymes. However, N2-domain seems to differ between from mostly bacterial and eukaryotic cells and in archaeal version. In fact for most archaeal cells and mitochodrial TARS, there is no presence of N2-domain and perform a different editing mechanism[15]. As mentioned earlier, AARS and TARS editing mechanism are homologous since the C-terminal domain of AARS and N2-domain of TARS both perform similar mechanism and use similar amino acids to select for serine bound tRNA(alanine or Thr) to hydrolyze and replace with correct amino acid[16].
Disease Relevance
Lately the aaRS family was found to have more function than just aminoacylation. For instance, many aaRs molecules have been found to link with angiogenesis, blood vessel growth occuring within a cancer environment[17]. This is seen in human exogenous TARS in its ability to generate blood vessels within ovarian cancer environment[18]. Studies on the structure of TARS bound to BC194, derivative to the natural antibiotic Borrelidin, were investigated to understand how the angiogenic signaling from TARS occurs[19].
List to available structures
Bacterial
E. coli: 1qf6Structure of E. coli TARS in the presence of tRNA
S. Aureus: 1nyq
Yeast (Saccharomyces cerevisiae): 3ugt
Eukaryotic
Human: 4hwt(w/ L-threonimide inhibitor); 4ttv (w/ BC194 inhibitor)
References
- ↑ Rajendran V, Kalita P, Shukla H, Kumar A, Tripathi T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int J Biol Macromol. 2018 May;111:400-414. doi: 10.1016/j.ijbiomac.2017.12.157., Epub 2018 Jan 3. PMID:29305884 doi:http://dx.doi.org/10.1016/j.ijbiomac.2017.12.157
- ↑ Rajendran V, Kalita P, Shukla H, Kumar A, Tripathi T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int J Biol Macromol. 2018 May;111:400-414. doi: 10.1016/j.ijbiomac.2017.12.157., Epub 2018 Jan 3. PMID:29305884 doi:http://dx.doi.org/10.1016/j.ijbiomac.2017.12.157
- ↑ Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). Lehninger principles of biochemistry. New York: Worth Publishers.
- ↑ Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). Lehninger principles of biochemistry. New York: Worth Publishers.
- ↑ Teng M, Hilgers MT, Cunningham ML, Borchardt A, Locke JB, Abraham S, Haley G, Kwan BP, Hall C, Hough GW, Shaw KJ, Finn J. Identification of bacteria-selective threonyl-tRNA synthetase substrate inhibitors by structure-based design. J Med Chem. 2013 Feb 28;56(4):1748-60. doi: 10.1021/jm301756m. Epub 2013 Feb 12. PMID:23362938 doi:10.1021/jm301756m
- ↑ Torres-Larios A, Sankaranarayanan R, Rees B, Dock-Bregeon AC, Moras D. Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase. J Mol Biol. 2003 Aug 1;331(1):201-11. PMID:12875846
- ↑ Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999 Aug;9(8):689-710. PMID:10447505
- ↑ http://www.rcsb.org/pdb/explore/macroMoleculeData.do?structureId=1TJE
- ↑ Dock-Bregeon AC, Rees B, Torres-Larios A, Bey G, Caillet J, Moras D. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004 Nov 5;16(3):375-86. PMID:15525511 doi:http://dx.doi.org/10.1016/j.molcel.2004.10.002
- ↑ Ling J, Peterson KM, Simonovic I, Soll D, Simonovic M. The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase. J Biol Chem. 2012 Aug 17;287(34):28518-25. doi: 10.1074/jbc.M112.372920. Epub, 2012 Jul 6. PMID:22773845 doi:http://dx.doi.org/10.1074/jbc.M112.372920
- ↑ Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999 Apr 30;97(3):371-81. PMID:10319817
- ↑ Minajigi A, Francklyn CS. RNA-assisted catalysis in a protein enzyme: The 2'-hydroxyl of tRNA(Thr) A76 promotes aminoacylation by threonyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2008 Nov 18;105(46):17748-53. doi:, 10.1073/pnas.0804247105. Epub 2008 Nov 7. PMID:18997014 doi:http://dx.doi.org/10.1073/pnas.0804247105
- ↑ Cura V, Moras D, Kern D. Sequence analysis and modular organization of threonyl-tRNA synthetase from Thermus thermophilus and its interrelation with threonyl-tRNA synthetases of other origins. Eur J Biochem. 2000 Jan;267(2):379-93. PMID:10632708
- ↑ Torres-Larios A, Sankaranarayanan R, Rees B, Dock-Bregeon AC, Moras D. Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase. J Mol Biol. 2003 Aug 1;331(1):201-11. PMID:12875846
- ↑ Beebe K, Merriman E, Ribas De Pouplana L, Schimmel P. A domain for editing by an archaebacterial tRNA synthetase. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):5958-63. doi:, 10.1073/pnas.0401530101. Epub 2004 Apr 12. PMID:15079065 doi:http://dx.doi.org/10.1073/pnas.0401530101
- ↑ Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999 Apr 30;97(3):371-81. PMID:10319817
- ↑ Mirando AC, Francklyn CS, Lounsbury KM. Regulation of angiogenesis by aminoacyl-tRNA synthetases. Int J Mol Sci. 2014 Dec 19;15(12):23725-48. doi: 10.3390/ijms151223725. PMID:25535072 doi:http://dx.doi.org/10.3390/ijms151223725
- ↑ Williams TF, Mirando AC, Wilkinson B, Francklyn CS, Lounsbury KM. Secreted Threonyl-tRNA synthetase stimulates endothelial cell migration and angiogenesis. Sci Rep. 2013;3:1317. doi: 10.1038/srep01317. PMID:23425968 doi:http://dx.doi.org/10.1038/srep01317
- ↑ Mirando AC, Fang P, Williams TF, Baldor LC, Howe AK, Ebert AM, Wilkinson B, Lounsbury KM, Guo M, Francklyn CS. Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor. Sci Rep. 2015 Aug 14;5:13160. doi: 10.1038/srep13160. PMID:26271225 doi:http://dx.doi.org/10.1038/srep13160
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.






