User:Jennifer Taylor/Sandbox 4
From Proteopedia
(Undo revision 2904670 by Jennifer Taylor (Talk)) |
|||
| (4 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
<StructureSection load='4q7q' size='340' side='right' caption='Structure of 4Q7Q' scene=''> | <StructureSection load='4q7q' size='340' side='right' caption='Structure of 4Q7Q' scene=''> | ||
| - | 4Q7Q is a protein found in ''Chitinophaga pinensis'', a soil bacterium in the sphingobacterial family. Its structure | + | 4Q7Q is a protein found in ''Chitinophaga pinensis'', a soil bacterium in the sphingobacterial family. Its structure was characterized by the 15-year Protein Structure Initiative (PSI) launched by the National Institutes of Health (NIH) and therefore exists in Protein Data Bank (PDB). Its function, however, has not. |
After structural and sequential analysis via various databases including BLAST, Pfam, Dali, PyMOL, and ProMOL, we initially predicted that 4Q7Q is a hydrolase. More specifically, we hypothesized that 4Q7Q is a lipase, an enzyme that can breaks down lipids to form fatty acids and a glycerol molecule. | After structural and sequential analysis via various databases including BLAST, Pfam, Dali, PyMOL, and ProMOL, we initially predicted that 4Q7Q is a hydrolase. More specifically, we hypothesized that 4Q7Q is a lipase, an enzyme that can breaks down lipids to form fatty acids and a glycerol molecule. | ||
| Line 23: | Line 23: | ||
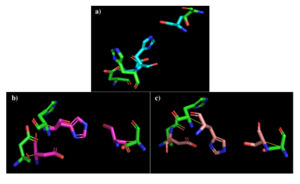
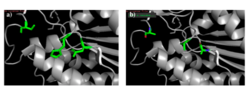
[[Image:alignments.png|thumb|right|300px|Figure 2: a) Alignment of 4Q7Q's putative catalytic triad (green) and 3LIP's catalytic triad (blue). b) Alignment of 4Q7Q's putative catalytic triad (green) and 1TAH's catalytic triad (pink). c) Alignment of 4Q7Q's putative catalytic triad (green) and 1BWR's catalytic triad (salmon).]] | [[Image:alignments.png|thumb|right|300px|Figure 2: a) Alignment of 4Q7Q's putative catalytic triad (green) and 3LIP's catalytic triad (blue). b) Alignment of 4Q7Q's putative catalytic triad (green) and 1TAH's catalytic triad (pink). c) Alignment of 4Q7Q's putative catalytic triad (green) and 1BWR's catalytic triad (salmon).]] | ||
| - | We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. The top hit was 4M8K, a GDSL-like lipase, a type of a lipase that has a flexible active site and therefore broad substrate specificity. Through BLAST, we found that 4M8K and 4Q7Q had a 36% sequence identity, with an E value of 0.002, indicating that it is a significant match. Since we can use the principle of homology to predict the function of an unknown protein, we first hypothesized that 4Q7Q was too a lipase. | + | We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. The top hit was 4M8K, a GDSL-like lipase, a type of a lipase that has a flexible active site and therefore broad substrate specificity. Through BLAST, we found that 4M8K and 4Q7Q had a 36% sequence identity, with an E value (a parameter that measures the number of alignments one can expect to see by chance) of 0.002, indicating that it is a significant match. Since we can use the principle of homology to predict the function of an unknown protein, we first hypothesized that 4Q7Q was too a lipase. |
| - | Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q is Ser164, Asp193, and His196. We believe that this group of amino acids are involved in the active site of 4Q7Q and therefore affects how the protein | + | Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q is Ser164, Asp193, and His196. We believe that this group of amino acids are involved in the active site of 4Q7Q and therefore affects how the protein performs. As seen in this <scene name='78/787192/4q7q_active_site/8'>image</scene>, all three amino acids are close in proximity to one another and are brought together in a single orientation. |
We also performed further analysis in PyMOL and ProMOL which involved the homology of active sites. Top hits included 3LIP, a lipase found in ''Burkholderia cepacia'' (a human pathogen that can cause pneumonia), 1TAH, a lipase found in ''Burkholderia glumae'' (a soil bacterium), and 1BWR, a hydrolase found in ''Bos taurus'' (cattle). We aligned the putative catalytic triad of 4Q7Q with each of the catalytic triads of these known proteins. | We also performed further analysis in PyMOL and ProMOL which involved the homology of active sites. Top hits included 3LIP, a lipase found in ''Burkholderia cepacia'' (a human pathogen that can cause pneumonia), 1TAH, a lipase found in ''Burkholderia glumae'' (a soil bacterium), and 1BWR, a hydrolase found in ''Bos taurus'' (cattle). We aligned the putative catalytic triad of 4Q7Q with each of the catalytic triads of these known proteins. | ||
| Line 35: | Line 35: | ||
1BWR has one chain. As seen in Figure 2c, when aligning the catalytic triad of 1BWR (Asp192, Ser47, His195) to the putative catalytic triad of 4Q7Q, the RMS is 2.049. | 1BWR has one chain. As seen in Figure 2c, when aligning the catalytic triad of 1BWR (Asp192, Ser47, His195) to the putative catalytic triad of 4Q7Q, the RMS is 2.049. | ||
| - | Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the lower RMS value measured. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. | + | Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the lower RMS (root mean square) value measured. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. |
| Line 65: | Line 65: | ||
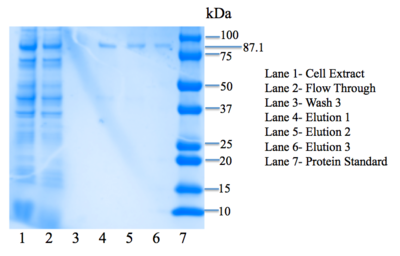
After the second bacterial transformation, BL21 cells were lysed and spread on LB+amp plates. A bacterial colony was then selected from a plate and suspended in liquid culture. After incubation overnight, the OD<sub>260</sub>, or the absorbance of the sample at 260 nm, was measured with a biophotometer. | After the second bacterial transformation, BL21 cells were lysed and spread on LB+amp plates. A bacterial colony was then selected from a plate and suspended in liquid culture. After incubation overnight, the OD<sub>260</sub>, or the absorbance of the sample at 260 nm, was measured with a biophotometer. | ||
| - | From our OD<sub>260</sub>, we calculated that our plasmid DNA concentration was 28.7 | + | From our OD<sub>260</sub>, we calculated that our plasmid DNA concentration was 28.7 μg/ mL. This concentration is low, probably due to the large size of our ORF. |
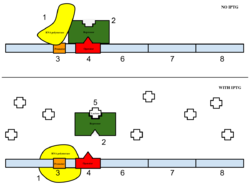
The sample was then induced with IPTG. As seen in Figure 4, IPTG is a reagent that prevents the repressor from binding to the operator to allow expression to occur. | The sample was then induced with IPTG. As seen in Figure 4, IPTG is a reagent that prevents the repressor from binding to the operator to allow expression to occur. | ||
| Line 107: | Line 107: | ||
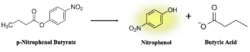
We performed a lipase activity assay using p-nitrophenyl butyrate. We added pNPB to 4Q7Q in an aqueous solution consisting of two solutions. One contained 50 mM Tris buffer and Triton-X, while the other contained pNPB dissolved in n-heptane. | We performed a lipase activity assay using p-nitrophenyl butyrate. We added pNPB to 4Q7Q in an aqueous solution consisting of two solutions. One contained 50 mM Tris buffer and Triton-X, while the other contained pNPB dissolved in n-heptane. | ||
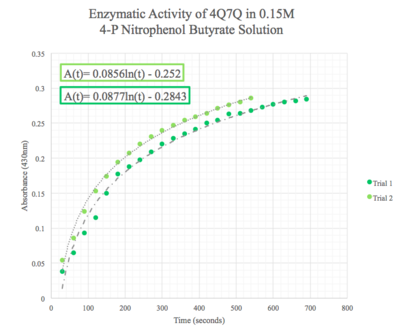
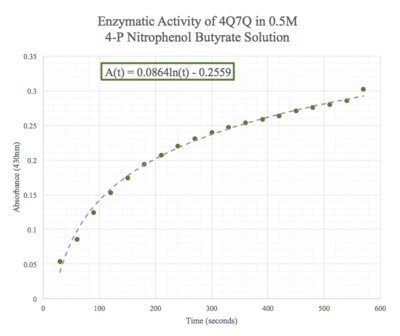
| - | Given the low concentration of our protein expressed during SDS-PAGE analysis, we employed a 10:1 ratio between the enzyme, 4Q7Q, and the substrate, pNPB. Molarities of 0.15 and 0.5 were tested for pNPB. | + | Given the low concentration of our protein expressed during SDS-PAGE analysis, we employed a 10:1 ratio between the enzyme, 4Q7Q, and the substrate, pNPB. Through increasing the ratio, we hoped that we would be able to observe 4Q7Q's enzymatic activity in a shorter period of time, given our time constraints in the lab. Molarities of 0.15 and 0.5 were tested for pNPB. |
A spectrophotometer was employed to measure the A<sub>430</sub> over a 10 minute period. | A spectrophotometer was employed to measure the A<sub>430</sub> over a 10 minute period. | ||
| Line 113: | Line 113: | ||
As seen in figure 7, a lipase breaks down pNPB into nitrophenol and butyric acid. Nitrophenol is yellow, therefore a color change can be detected. The spectrophotometer quantitatively measures the absorbance of the sample over time; we expect an increasing rate of nitrophenol production over time. When pNPB runs out, the rate should flatten out, as there will be no more reactants to turn into products. | As seen in figure 7, a lipase breaks down pNPB into nitrophenol and butyric acid. Nitrophenol is yellow, therefore a color change can be detected. The spectrophotometer quantitatively measures the absorbance of the sample over time; we expect an increasing rate of nitrophenol production over time. When pNPB runs out, the rate should flatten out, as there will be no more reactants to turn into products. | ||
| - | We then plotted the data of A<sub>430</sub> vs. time in seconds. | + | We then plotted the data of A<sub>430</sub> vs. time in seconds. We can see that nitrophenol was being produced at an increasing rate and therefore conclude that 4Q7Q acts like a lipase. |
[[Image:0.15M_pNPB.png|thumb|left|400px|Figure 8: Enzymatic activity of 4Q7Q in 0.15 M pNPB. The ratio of 4Q7Q to pNPB is 10:1.]] | [[Image:0.15M_pNPB.png|thumb|left|400px|Figure 8: Enzymatic activity of 4Q7Q in 0.15 M pNPB. The ratio of 4Q7Q to pNPB is 10:1.]] | ||
| Line 161: | Line 161: | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
1) PDB ID: 4Q7Q. Tan, K., Tesar, C., Clancy, S., Joachimiak. A., The crystal structure of a possible lipase from Chitinophaga pinesis DSM 2588. | 1) PDB ID: 4Q7Q. Tan, K., Tesar, C., Clancy, S., Joachimiak. A., The crystal structure of a possible lipase from Chitinophaga pinesis DSM 2588. | ||
| Line 177: | Line 178: | ||
8) Zhong, Q., Glatz, C. (2006) Enzymatic Assay Method for Evaluating the Lipase Activity in Complex Extracts from Transgenic Corn Seed. | 8) Zhong, Q., Glatz, C. (2006) Enzymatic Assay Method for Evaluating the Lipase Activity in Complex Extracts from Transgenic Corn Seed. | ||
| + | |||
| + | 9) Alberts, Bruce. Molecular Biology of the Cell. 6th ed. New York, NY: Garland Science, Taylor and Francis Group, 2015. | ||
| + | |||
| + | ==Author== | ||
| + | |||
| + | Julia Angkeow | ||
Current revision
4Q7Q
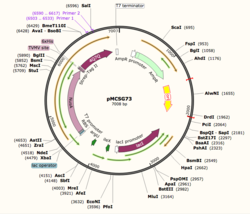
| |||||||||||
References
1) PDB ID: 4Q7Q. Tan, K., Tesar, C., Clancy, S., Joachimiak. A., The crystal structure of a possible lipase from Chitinophaga pinesis DSM 2588.
2) Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410/
3) Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., Salazar G.A., Tate J., Bateman A., The Pfam protein families database: towards a more sustainable future.
4) Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucl. Acids Res. 38, W545-549.
5) SnapGene software (from GSL Biotech; available at snapgene.com).
6) The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
7) Craig P., Bernstein, H., Mills J., ProMOL software (from Rochester Institute of Technology, available at promol.org).
8) Zhong, Q., Glatz, C. (2006) Enzymatic Assay Method for Evaluating the Lipase Activity in Complex Extracts from Transgenic Corn Seed.
9) Alberts, Bruce. Molecular Biology of the Cell. 6th ed. New York, NY: Garland Science, Taylor and Francis Group, 2015.
Author
Julia Angkeow