We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Jennifer Taylor/Sandbox 1
From Proteopedia
< User:Jennifer Taylor(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 11: | Line 11: | ||
A common theme in biology is that form follows function. Thus, we used computer programs to find which proteins were most homologous to YxiM in terms of sequence and structure, with the expectation that YxiM is likely to be functionally similar to those proteins that have similar sequences and structures. | A common theme in biology is that form follows function. Thus, we used computer programs to find which proteins were most homologous to YxiM in terms of sequence and structure, with the expectation that YxiM is likely to be functionally similar to those proteins that have similar sequences and structures. | ||
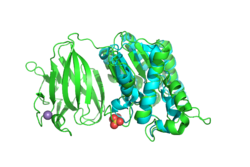
| - | + | [[Image:YxiM_green-1J00_blue.png|thumb|left|250px|'''Figure 1'''. YxiM (green) aligned with 1J00 (blue), RMSD = 2.692.]] | |
| - | + | We used BLAST and PFam to find characterized proteins whose sequences aligned best with YxiM. Sequence analysis suggests that YxiM is a GDSL-like lipase, a type of esterase. Esterases are molecules that hydrolyze (decompose) a class of organic molecules known as esters. GDSL-like lipases demonstrate broad substrate specificity due to their flexible structures. BLAST showed that the proteins 1J00, 1IVN, and 1JRL have the highest sequence homology to YxiM. These proteins are multifunctional hydrolases that show both esterase and protease activity. | |
Next, we used PyMOL to align the 3D structures of the BLAST hits with that of YxiM. The proteins 1J00, 1IVN, and 1JRL all align well with the α-helix domain of YxiM. | Next, we used PyMOL to align the 3D structures of the BLAST hits with that of YxiM. The proteins 1J00, 1IVN, and 1JRL all align well with the α-helix domain of YxiM. | ||
| + | |||
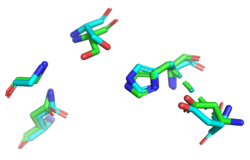
| + | [[Image:YxiM-1BWR_Catalytic.png|thumb|right|250px|'''Figure 2'''. Active site of 1BWR aligned with the putative active site of YxiM, RMSD = 0.506.]] | ||
The Dali server finds the most similar proteins based on 3D structures, and the top 30 hits for YxiM were are all rhamnogalacturonan acetylesterases, GDSL lipases, LAE5s (hydrolases), or acetyl xylan esterases, which further suggests that YxiM is an esterase. | The Dali server finds the most similar proteins based on 3D structures, and the top 30 hits for YxiM were are all rhamnogalacturonan acetylesterases, GDSL lipases, LAE5s (hydrolases), or acetyl xylan esterases, which further suggests that YxiM is an esterase. | ||
Finally, we used ProMOL to perform a structural alignment of active sites of other proteins with YxiM to predict the active site of YxiM. We found that YxiM aligns best with the active site of IBWR, which is an esterase. The <scene name='78/787191/2o14_active_site/2'>putative catalytic triad</scene> of YxiM consists of amino acids S171, D339, and H342. | Finally, we used ProMOL to perform a structural alignment of active sites of other proteins with YxiM to predict the active site of YxiM. We found that YxiM aligns best with the active site of IBWR, which is an esterase. The <scene name='78/787191/2o14_active_site/2'>putative catalytic triad</scene> of YxiM consists of amino acids S171, D339, and H342. | ||
| - | |||
| - | [[Image:YxiM-1BWR_Catalytic.png|thumb|right|250px|Figure 2: insert your caption]] | ||
Based on these analyses, we predicted that YxiM is an esterase and proceeded to perform ''in vitro'' assays to confirm esterase activity. | Based on these analyses, we predicted that YxiM is an esterase and proceeded to perform ''in vitro'' assays to confirm esterase activity. | ||
| Line 27: | Line 27: | ||
== Plasmid Purification == | == Plasmid Purification == | ||
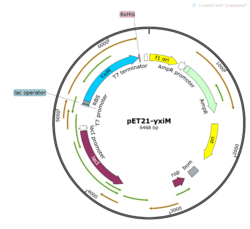
| - | + | [[Image:PET21-YxiM_Map.png|thumb|right|250px|'''Figure 3'''. Plasmid map of pET21-''yxiM''.]] | |
| - | + | In order to study the protein YxiM, we ordered a plasmid that contains the gene that transcribes the protein. A plasmid is a type of circular bacterial DNA. By transforming (inserting) this plasmid (pET21-''yxiM'') into the bacteria (DH5α Competent ''E. coli''), we can use the bacteria to create more of the plasmid. Then, we performed a DNA miniprep to purify the plasmid for later use. | |
== Bacterial Transformation == | == Bacterial Transformation == | ||
| Line 44: | Line 44: | ||
== Esterase Activity Assay == | == Esterase Activity Assay == | ||
| + | |||
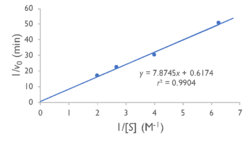
| + | [[Image:YxiM_Lineweaver-Burk_Plot.png|thumb|left|250px|'''Figure 4'''. Lineweaver-Burk plot of YxiM esterase activity.]] | ||
Now that we had purified protein, we could test the function of YxiM ''in vitro''. Since we believed that YxiM was an ester, we placed it in a buffered solution with 4-nitrophenyl butyrate, a type of ester. Esterases should hydrolyze 4-nitrophenyl butyrate, causing the products butyric acid and 4-nitrophenol to form. Since 4-nitrophenyl is a yellow color, the absorbance of the solution changes as more products are formed. We used colorimeter to measure the absorbance at 430 nm as a proxy for esterase activity. We found that the absorbance increases over time, which suggests that YxiM is indeed an esterase. | Now that we had purified protein, we could test the function of YxiM ''in vitro''. Since we believed that YxiM was an ester, we placed it in a buffered solution with 4-nitrophenyl butyrate, a type of ester. Esterases should hydrolyze 4-nitrophenyl butyrate, causing the products butyric acid and 4-nitrophenol to form. Since 4-nitrophenyl is a yellow color, the absorbance of the solution changes as more products are formed. We used colorimeter to measure the absorbance at 430 nm as a proxy for esterase activity. We found that the absorbance increases over time, which suggests that YxiM is indeed an esterase. | ||
Specifically, we found that the Lineweaver-Burk plot of esterase activity is linear. This is typical of enzymes, as predicted by the Michaelis-Menten model of enzyme kinetics. Technically, we did not construct a true Lineweaver-Burk plot, as we used absorbance as a proxy for molar concentration, but absorbance varies linearly with concentration, as shown by the Beer-Lambert law. | Specifically, we found that the Lineweaver-Burk plot of esterase activity is linear. This is typical of enzymes, as predicted by the Michaelis-Menten model of enzyme kinetics. Technically, we did not construct a true Lineweaver-Burk plot, as we used absorbance as a proxy for molar concentration, but absorbance varies linearly with concentration, as shown by the Beer-Lambert law. | ||
| - | |||
| - | [[Image:YxiM_Lineweaver-Burk_Plot.png|thumb|left|250px|Figure 2: insert your caption]] | ||
== Discussion == | == Discussion == | ||
Current revision
YxiM from Bacillus subtilis
| |||||||||||