User:Jennifer Taylor/Sandbox 7
From Proteopedia
< User:Jennifer Taylor(Difference between revisions)
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
==About 4Q7Q:A Hydrolase== | ==About 4Q7Q:A Hydrolase== | ||
| - | Here is a polar hydrophobic residue image of my protein, <scene name='78/787195/4q7q_polar_and_hydrophobic/1'>4Q7Q</scene>. | ||
<StructureSection load='4q7q' size='340' side='both' caption='Caption for this structure' scene=''> | <StructureSection load='4q7q' size='340' side='both' caption='Caption for this structure' scene=''> | ||
This is a default text for your page '''Jennifer Taylor/Sandbox 7'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Jennifer Taylor/Sandbox 7'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| Line 6: | Line 5: | ||
==What is 4Q7Q?== | ==What is 4Q7Q?== | ||
| - | In 2000, the protein structure initiative (PSI) began with the goal of finding the three dimensional structure of as many proteins as possible. In 2010, however, funding for the PSI was terminated. Nevertheless, millions of protein structures were found, and many structures were uncharacterized. One of these uncharacterized proteins was <scene name='78/787195/4q7q_assymetric_assembly/1'>4Q7Q</scene>, which is found in <i>Chitinophaga pinesis</i>. Its molecular weight is 87.1 kDa. Through structural and sequential analysis on BLAST, PFam, Dali, and ProMol, 4Q7Q is believed to be a lipase. | + | In 2000, the protein structure initiative (PSI) began with the goal of finding the three-dimensional structure of as many proteins as possible. In 2010, however, funding for the PSI was terminated. Nevertheless, millions of protein structures were found, and many structures were uncharacterized. One of these uncharacterized proteins was <scene name='78/787195/4q7q_assymetric_assembly/1'>4Q7Q</scene>, which is found in <i>Chitinophaga pinesis</i>. Its molecular weight is 87.1 kDa. Through structural and sequential analysis performed on BLAST, PFam, Dali, and ProMol, 4Q7Q is believed to be a lipase. |
==Sequential analysis of 4Q7Q== | ==Sequential analysis of 4Q7Q== | ||
| - | <b>BLAST</b>-Sequence homology between 4Q7Q was tested through BLAST; the top hit for 4Q7Q was 4M8K, a GDSL-like lipase. 4Q7Q and 4M8k share a 36% sequence homology, but a low E-value, indicating it is a significant match. | + | <b>BLAST</b>-Sequence homology between 4Q7Q was tested through BLAST; the top hit for 4Q7Q was <scene name='78/787195/4m8k/1'>4M8K</scene>, a GDSL-like lipase. 4Q7Q and 4M8k share a 36% sequence homology, but a low E-value, indicating it is a significant structural match of 4Q7Q. |
<b>PFam</b>-Results from PFam sequence mapping also indicated that 4Q7Q shares sequence homology with other GDSL-like lipases. | <b>PFam</b>-Results from PFam sequence mapping also indicated that 4Q7Q shares sequence homology with other GDSL-like lipases. | ||
| Line 18: | Line 17: | ||
<b>Dali</b>-Top hits from Dali indicated that 4Q7Q shares structural homology with putative lipases. | <b>Dali</b>-Top hits from Dali indicated that 4Q7Q shares structural homology with putative lipases. | ||
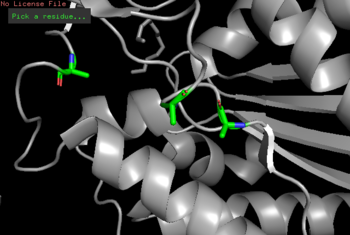
| - | <b>ProMol</b>-Through ProMol, a possible catalytic triad for 4Q7Q (Ser164, Asp193, His196) was found due to catalytic triad alignment with 1BRW. 4Q7Q and 1BRW catalytic triad alignment yielded | + | <b>ProMol</b>-Through ProMol, a possible catalytic triad for <scene name='78/787195/4q7q_catalytic_triad/1'>4Q7Q</scene> (Ser164, Asp193, His196) was found due to catalytic triad alignment with 1BRW. 4Q7Q and 1BRW catalytic triad alignment yielded an RMS value of 2.049 and a full structural homology with an RMS value of 4.852. |
==Enzymatic Assay of 4Q7Q== | ==Enzymatic Assay of 4Q7Q== | ||
| - | <b>Reasoning</b>-Results obtained in silico strongly suggested that 4Q7Q was most likely to be hydrolase | + | <b>Reasoning</b>-Results obtained in silico strongly suggested that 4Q7Q was most likely to be hydrolase (and specifically a lipase). Thus, a lipase assay was run on 4Q7Q to confirm its enzymatic function. The lipase assay conducted was done through spectrophotometric methods. Purified 4Q7Q was mixed in solution with P-nitrophenyl butyrate. When nitrophenyl butyrate undergoes hydrolysis, its products are p-nitrophenol and butyrate. P-nitrophenol’s color becomes yellow when this reaction happens in solution. Thus, when a lipase catalyzes the hydrolysis of P-nitrophenol butyrate, a color change in solution will occur. A colorimeter was set at 430nm to detect light in the “yellow” range and thereby measure the change in color occurring when 4Q7Q catalyzed the hydrolysis reaction. |
<b>4.2 Results</b> | <b>4.2 Results</b> | ||
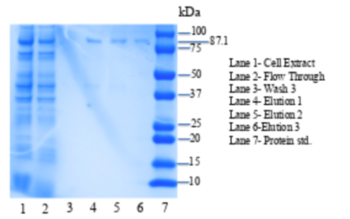
| + | [[Image:4q7q_SDS.png|thumb|left|350px|Figure 2: SDS-PAGE gel of purified 4Q7Q plasmid. L1: Cell Extract, L2:Flow Through,L3: Wash 3, L4: Elution 1 , L5: Elution 2, L6: Elution 3 , L7: Protein standards.]] {{clear}} | ||
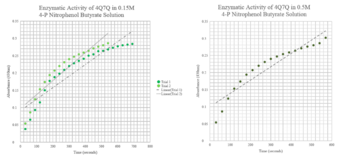
| - | <b>Conclusion</b>-Due to 4Q7Q’s low concentration found on the SDS-page gel, 4Q7Q to pNPB ratio in solution was 10:1. A logarithmic | + | [[Image:pNPB assay.png|thumb|left|350px|Figure 3:pNPB Assay Results for 4Q7Q.]] {{clear}} |
| + | |||
| + | <b>Conclusion</b>-Due to 4Q7Q’s low concentration found on the SDS-page gel, 4Q7Q to p-Nitrophenol Butyrate (pNPB) ratio in solution was 10:1. A pattern with a logarithmic character was observed during the lipase assay conducted on 4Q7Q, indicating a successful catalysis of a lipid through hydrolysis. | ||
==Future directions== | ==Future directions== | ||
| - | <b>Cloning</b>-To confirm the suspected catalytic triad of 4Q7Q, site directed mutagenesis of 4Q7Qs catalytic triad was performed through PCR. Substitutions of residues were completed as followed: Ser164Ala, Asp193Ala, His196Ala. Following a successful cloning, a p-NPB lipase assay will be performed to see whether the new mutations on 4Q7Q deterred catalysis of the hydrolysis reaction of p-NPB. | + | <b>Cloning</b>-To confirm the suspected catalytic triad of 4Q7Q as suggested by ProMol analysis, site-directed mutagenesis of 4Q7Qs catalytic triad was performed through PCR. Substitutions of residues were completed as followed: Ser164Ala, Asp193Ala, His196Ala. Following a successful cloning, a p-NPB lipase assay will be performed to see whether the new mutations on 4Q7Q deterred catalysis of the hydrolysis reaction of p-NPB. A deterred would indicate the cloned sequences did, in fact, mutate the catalytic triad, altering the structure of the cloned 4Q7Q and affecting enzymatic activity. |
| - | + | [[Image:4Q7Q_Mutagenesis.png|thumb|left|350px|Figure 3:Mutagenesis of 4Q7Q (Ser164Ala, Asp193Ala, His196Ala).]] {{clear}} | |
| - | + | <b>Enzyme Kinetics of 4Q7Q</b>=Further data of enzyme activity of 4Q7Q using our lipase assay at different concentrations of p-NPB can be performed to create a Lineweaver-Burk plot that would deepen analysis and understanding of the enzymatic activity of 4Q7Q. | |
| + | <b>Optimization of enzymatic activity</b>-Conditions for optimized enzyme activity can be tested by performing iterations of the p-NPB assay by altering environmental conditions of the enzyme (pH and temperature). | ||
Current revision
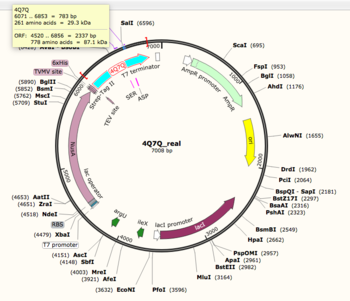
About 4Q7Q:A Hydrolase
| |||||||||||
References and Notes
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644